| 1 | d1v4va_ |
|
 |
100.0 |
15 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
| 2 | d1o6ca_ |
|
 |
100.0 |
15 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
| 3 | c3ot5D_ |
|
 |
100.0 |
13 |
PDB header:isomerase
Chain: D: PDB Molecule:udp-n-acetylglucosamine 2-epimerase;
PDBTitle: 2.2 angstrom resolution crystal structure of putative udp-n-2 acetylglucosamine 2-epimerase from listeria monocytogenes
|
| 4 | d1f6da_ |
|
 |
100.0 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
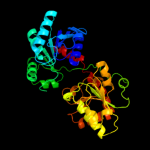
| 5 | c3c4vB_ |
|
 |
100.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:predicted glycosyltransferases;
PDBTitle: structure of the retaining glycosyltransferase msha:the2 first step in mycothiol biosynthesis. organism:3 corynebacterium glutamicum : complex with udp and 1l-ins-1-4 p.
|
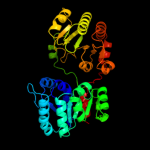
| 6 | c3dzcA_ |
|
 |
100.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 2-epimerase;
PDBTitle: 2.35 angstrom resolution structure of wecb (vc0917), a udp-n-2 acetylglucosamine 2-epimerase from vibrio cholerae.
|
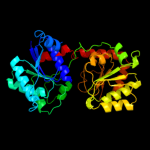
| 7 | c2jjmH_ |
|
 |
100.0 |
14 |
PDB header:transferase
Chain: H: PDB Molecule:glycosyl transferase, group 1 family protein;
PDBTitle: crystal structure of a family gt4 glycosyltransferase from2 bacillus anthracis orf ba1558.
|
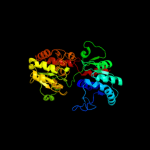
| 8 | c3s29C_ |
|
 |
100.0 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:sucrose synthase 1;
PDBTitle: the crystal structure of sucrose synthase-1 from arabidopsis thaliana2 and its functional implications.
|
| 9 | c2qzsA_ |
|
 |
100.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:glycogen synthase;
PDBTitle: crystal structure of wild-type e.coli gs in complex with adp2 and glucose(wtgsb)
|
| 10 | c2r60A_ |
|
 |
100.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:glycosyl transferase, group 1;
PDBTitle: structure of apo sucrose phosphate synthase (sps) of2 halothermothrix orenii
|
| 11 | c2gejA_ |
|
 |
100.0 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol mannosyltransferase (pima);
PDBTitle: crystal structure of phosphatidylinositol mannosyltransferase (pima)2 from mycobacterium smegmatis in complex with gdp-man
|
| 12 | c3okaA_ |
|
 |
100.0 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:gdp-mannose-dependent alpha-(1-6)-phosphatidylinositol
PDBTitle: crystal structure of corynebacterium glutamicum pimb' in complex with2 gdp-man (triclinic crystal form)
|
| 13 | d2bisa1 |
|
 |
100.0 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 14 | c2x6rA_ |
|
 |
100.0 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:trehalose-synthase tret;
PDBTitle: crystal structure of trehalose synthase tret from p.2 horikoshi produced by soaking in trehalose
|
| 15 | d1rzua_ |
|
 |
100.0 |
12 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 16 | d1f0ka_ |
|
 |
99.9 |
13 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Peptidoglycan biosynthesis glycosyltransferase MurG |
| 17 | c2xmpB_ |
|
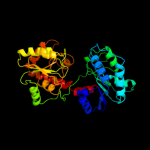 |
99.9 |
13 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:trehalose-synthase tret;
PDBTitle: crystal structure of trehalose synthase tret mutant e326a2 from p.horishiki in complex with udp
|
| 18 | d2iw1a1 |
|
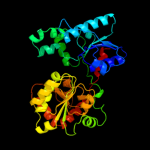 |
99.9 |
13 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 19 | c3oy2A_ |
|
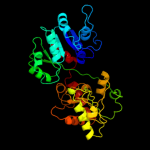 |
99.9 |
11 |
PDB header:viral protein,transferase
Chain: A: PDB Molecule:glycosyltransferase b736l;
PDBTitle: crystal structure of a putative glycosyltransferase from paramecium2 bursaria chlorella virus ny2a
|
| 20 | c1uquB_ |
|
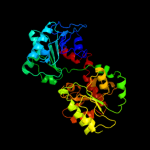 |
99.9 |
14 |
PDB header:synthase
Chain: B: PDB Molecule:alpha, alpha-trehalose-phosphate synthase;
PDBTitle: trehalose-6-phosphate from e. coli bound with udp-glucose.
|
| 21 | c2xcuC_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:3-deoxy-d-manno-2-octulosonic acid transferase;
PDBTitle: membrane-embedded monofunctional glycosyltransferase waaa of aquifex2 aeolicus, comlex with cmp
|
| 22 | c3ia7A_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:calg4;
PDBTitle: crystal structure of calg4, the calicheamicin glycosyltransferase
|
| 23 | c2iyaB_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:oleandomycin glycosyltransferase;
PDBTitle: the crystal structure of macrolide glycosyltransferases: a2 blueprint for antibiotic engineering
|
| 24 | d1uqta_ |
|
not modelled |
99.9 |
13 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Trehalose-6-phosphate synthase, OtsA |
| 25 | c3iaaB_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:calg2;
PDBTitle: crystal structure of calg2, calicheamicin glycosyltransferase, tdp2 bound form
|
| 26 | c2iyfA_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:oleandomycin glycosyltransferase;
PDBTitle: the crystal structure of macrolide glycosyltransferases: a2 blueprint for antibiotic engineering
|
| 27 | c3nb0A_ |
|
not modelled |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:glycogen [starch] synthase isoform 2;
PDBTitle: glucose-6-phosphate activated form of yeast glycogen synthase
|
| 28 | c3o3cD_ |
|
not modelled |
99.9 |
12 |
PDB header:transferase
Chain: D: PDB Molecule:glycogen [starch] synthase isoform 2;
PDBTitle: glycogen synthase basal state udp complex
|
| 29 | c3othB_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase/antibiotic
Chain: B: PDB Molecule:calg1;
PDBTitle: crystal structure of calg1, calicheamicin glycostyltransferase, tdp2 and calicheamicin alpha3i bound form
|
| 30 | c2x0dA_ |
|
not modelled |
99.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:wsaf;
PDBTitle: apo structure of wsaf
|
| 31 | c2p6pB_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyl transferase;
PDBTitle: x-ray crystal structure of c-c bond-forming dtdp-d-olivose-transferase2 urdgt2
|
| 32 | c2q6vA_ |
|
not modelled |
99.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:glucuronosyltransferase gumk;
PDBTitle: crystal structure of gumk in complex with udp
|
| 33 | c3d0qB_ |
|
not modelled |
99.8 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:protein calg3;
PDBTitle: crystal structure of calg3 from micromonospora echinospora determined2 in space group i222
|
| 34 | d1pn3a_ |
|
not modelled |
99.8 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Gtf glycosyltransferase |
| 35 | d1rrva_ |
|
not modelled |
99.8 |
15 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Gtf glycosyltransferase |
| 36 | d1iira_ |
|
not modelled |
99.8 |
11 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Gtf glycosyltransferase |
| 37 | c2iv3B_ |
|
not modelled |
99.8 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyltransferase;
PDBTitle: crystal structure of avigt4, a glycosyltransferase involved2 in avilamycin a biosynthesis
|
| 38 | c3hbjA_ |
|
not modelled |
99.8 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:flavonoid 3-o-glucosyltransferase;
PDBTitle: structure of ugt78g1 complexed with udp
|
| 39 | c3rhzB_ |
|
not modelled |
99.7 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:nucleotide sugar synthetase-like protein;
PDBTitle: structure and functional analysis of a new subfamily of2 glycosyltransferases required for glycosylation of serine-rich3 streptococcal adhesions
|
| 40 | c2vsnB_ |
|
not modelled |
99.7 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:xcogt;
PDBTitle: structure and topological arrangement of an o-glcnac2 transferase homolog: insight into molecular control of3 intracellular glycosylation
|
| 41 | d2acva1 |
|
not modelled |
99.7 |
8 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 42 | d2c1xa1 |
|
not modelled |
99.7 |
11 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 43 | d2pq6a1 |
|
not modelled |
99.6 |
13 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 44 | c3qhpB_ |
|
not modelled |
99.5 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:type 1 capsular polysaccharide biosynthesis protein j
PDBTitle: crystal structure of the catalytic domain of cholesterol-alpha-2 glucosyltransferase from helicobacter pylori
|
| 45 | d2vcha1 |
|
not modelled |
99.5 |
12 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 46 | c3pe3D_ |
|
not modelled |
99.4 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:udp-n-acetylglucosamine--peptide n-
PDBTitle: structure of human o-glcnac transferase and its complex with a peptide2 substrate
|
| 47 | c3hbmA_ |
|
not modelled |
99.4 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:udp-sugar hydrolase;
PDBTitle: crystal structure of pseg from campylobacter jejuni
|
| 48 | d2bfwa1 |
|
not modelled |
99.4 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 49 | d2f9fa1 |
|
not modelled |
99.3 |
10 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 50 | c3tovB_ |
|
not modelled |
99.0 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyl transferase family 9;
PDBTitle: the crystal structure of the glycosyl transferase family 9 from2 veillonella parvula dsm 2008
|
| 51 | c2h1fB_ |
|
not modelled |
98.7 |
7 |
PDB header:transferase
Chain: B: PDB Molecule:lipopolysaccharide heptosyltransferase-1;
PDBTitle: e. coli heptosyltransferase waac with adp
|
| 52 | c3q3hA_ |
|
not modelled |
98.7 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:hmw1c-like glycosyltransferase;
PDBTitle: crystal structure of the actinobacillus pleuropneumoniae hmw1c2 glycosyltransferase in complex with udp-glc
|
| 53 | d1pswa_ |
|
not modelled |
98.7 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:ADP-heptose LPS heptosyltransferase II |
| 54 | c2o6lA_ |
|
not modelled |
98.4 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:udp-glucuronosyltransferase 2b7;
PDBTitle: crystal structure of the udp-glucuronic acid binding domain2 of the human drug metabolizing udp-glucuronosyltransferase3 2b7
|
| 55 | c3l7mC_ |
|
not modelled |
98.4 |
13 |
PDB header:structural protein
Chain: C: PDB Molecule:teichoic acid biosynthesis protein f;
PDBTitle: structure of the wall teichoic acid polymerase tagf, h548a
|
| 56 | c2jzcA_ |
|
not modelled |
96.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine transferase subunit
PDBTitle: nmr solution structure of alg13: the sugar donor subunit of2 a yeast n-acetylglucosamine transferase. northeast3 structural genomics consortium target yg1
|
| 57 | c2pzlB_ |
|
not modelled |
95.4 |
22 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme2 wbmg in complex with nad and udp
|
| 58 | c3oh8A_ |
|
not modelled |
95.2 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate sugar epimerase (sula family);
PDBTitle: crystal structure of the nucleoside-diphosphate sugar epimerase from2 corynebacterium glutamicum. northeast structural genomics consortium3 target cgr91
|
| 59 | d2c5aa1 |
|
not modelled |
94.8 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 60 | c2hunB_ |
|
not modelled |
94.7 |
26 |
PDB header:lyase
Chain: B: PDB Molecule:336aa long hypothetical dtdp-glucose 4,6-dehydratase;
PDBTitle: crystal structure of hypothetical protein ph0414 from pyrococcus2 horikoshii ot3
|
| 61 | c1zh8B_ |
|
not modelled |
94.4 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
| 62 | c2nvwB_ |
|
not modelled |
94.3 |
9 |
PDB header:transcription
Chain: B: PDB Molecule:galactose/lactose metabolism regulatory protein
PDBTitle: crystal sctucture of transcriptional regulator gal80p from2 kluyveromymes lactis
|
| 63 | c3l77A_ |
|
not modelled |
94.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain alcohol dehydrogenase;
PDBTitle: x-ray structure alcohol dehydrogenase from archaeon thermococcus2 sibiricus complexed with 5-hydroxy-nadp
|
| 64 | d1ydga_ |
|
not modelled |
93.9 |
21 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
| 65 | c3zquA_ |
|
not modelled |
93.8 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:probable aromatic acid decarboxylase;
PDBTitle: structure of a probable aromatic acid decarboxylase
|
| 66 | c3e5nA_ |
|
not modelled |
93.7 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine-d-alanine ligase a;
PDBTitle: crystal strucutre of d-alanine-d-alanine ligase from2 xanthomonas oryzae pv. oryzae kacc10331
|
| 67 | d1fjha_ |
|
not modelled |
93.3 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 68 | c3q2kB_ |
|
not modelled |
92.8 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
|
| 69 | d1zh8a1 |
|
not modelled |
92.4 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 70 | d1mkza_ |
|
not modelled |
92.4 |
23 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 71 | c3lwbA_ |
|
not modelled |
92.2 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: crystal structure of apo d-alanine:d-alanine ligase (ddl) from2 mycobacterium tuberculosis
|
| 72 | c3tqrA_ |
|
not modelled |
92.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: structure of the phosphoribosylglycinamide formyltransferase (purn) in2 complex with ches from coxiella burnetii
|
| 73 | d1e7wa_ |
|
not modelled |
92.0 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 74 | d3c96a1 |
|
not modelled |
91.5 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 75 | c3enkB_ |
|
not modelled |
91.5 |
29 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: 1.9a crystal structure of udp-glucose 4-epimerase from2 burkholderia pseudomallei
|
| 76 | c3allA_ |
|
not modelled |
91.2 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;
PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase, mutant y270a
|
| 77 | c2qhxB_ |
|
not modelled |
91.2 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pteridine reductase 1;
PDBTitle: structure of pteridine reductase from leishmania major2 complexed with a ligand
|
| 78 | c3tqtB_ |
|
not modelled |
91.1 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine--d-alanine ligase;
PDBTitle: structure of the d-alanine-d-alanine ligase from coxiella burnetii
|
| 79 | c3i4fD_ |
|
not modelled |
90.9 |
22 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-oxoacyl-[acyl-carrier protein] reductase;
PDBTitle: structure of putative 3-oxoacyl-reductase from bacillus thuringiensis
|
| 80 | c3icpA_ |
|
not modelled |
90.9 |
30 |
PDB header:isomerase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of udp-galactose 4-epimerase
|
| 81 | c2p5uC_ |
|
not modelled |
90.9 |
20 |
PDB header:isomerase
Chain: C: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-2 epimerase complex with nad
|
| 82 | c3m2pD_ |
|
not modelled |
90.6 |
29 |
PDB header:isomerase
Chain: D: PDB Molecule:udp-n-acetylglucosamine 4-epimerase;
PDBTitle: the crystal structure of udp-n-acetylglucosamine 4-epimerase2 from bacillus cereus
|
| 83 | c2pvpB_ |
|
not modelled |
90.6 |
23 |
PDB header:ligase
Chain: B: PDB Molecule:d-alanine-d-alanine ligase;
PDBTitle: crystal structure of d-alanine-d-alanine ligase from helicobacter2 pylori
|
| 84 | d1wmaa1 |
|
not modelled |
90.6 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 85 | c1qzuB_ |
|
not modelled |
90.5 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:hypothetical protein mds018;
PDBTitle: crystal structure of human phosphopantothenoylcysteine decarboxylase
|
| 86 | d2ag5a1 |
|
not modelled |
90.4 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 87 | c3se7A_ |
|
not modelled |
90.2 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:vana;
PDBTitle: ancient vana
|
| 88 | d1u7za_ |
|
not modelled |
90.1 |
26 |
Fold:Ribokinase-like
Superfamily:CoaB-like
Family:CoaB-like |
| 89 | c1fmtA_ |
|
not modelled |
90.1 |
20 |
PDB header:formyltransferase
Chain: A: PDB Molecule:methionyl-trna fmet formyltransferase;
PDBTitle: methionyl-trnafmet formyltransferase from escherichia coli
|
| 90 | c3tfoD_ |
|
not modelled |
90.0 |
23 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative 3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of a putative 3-oxoacyl-(acyl-carrier-protein)2 reductase from sinorhizobium meliloti
|
| 91 | c3p19A_ |
|
not modelled |
89.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative blue fluorescent protein;
PDBTitle: improved nadph-dependent blue fluorescent protein
|
| 92 | c3ceaA_ |
|
not modelled |
89.8 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
| 93 | d1jaya_ |
|
not modelled |
89.7 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 94 | d1kewa_ |
|
not modelled |
89.6 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 95 | c2jahB_ |
|
not modelled |
89.6 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:clavulanic acid dehydrogenase;
PDBTitle: biochemical and structural analysis of the clavulanic acid2 dehydeogenase (cad) from streptomyces clavuligerus
|
| 96 | c2x4gA_ |
|
not modelled |
89.6 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate-sugar epimerase;
PDBTitle: crystal structure of pa4631, a nucleoside-diphosphate-sugar2 epimerase from pseudomonas aeruginosa
|
| 97 | c1zx9A_ |
|
not modelled |
89.5 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mercuric reductase;
PDBTitle: crystal structure of tn501 mera
|
| 98 | c3i1jB_ |
|
not modelled |
89.5 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, short chain
PDBTitle: structure of a putative short chain dehydrogenase from2 pseudomonas syringae
|
| 99 | d1fmta2 |
|
not modelled |
89.4 |
17 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 100 | d1udca_ |
|
not modelled |
89.4 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 101 | c3l6dB_ |
|
not modelled |
89.3 |
7 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from pseudomonas putida2 kt2440
|
| 102 | c3rkuC_ |
|
not modelled |
89.2 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase ymr226c;
PDBTitle: substrate fingerprint and the structure of nadp+ dependent serine2 dehydrogenase from saccharomyces cerevisiae complexed with nadp+
|
| 103 | d1b74a1 |
|
not modelled |
89.1 |
13 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 104 | c3edmD_ |
|
not modelled |
89.0 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase from agrobacterium2 tumefaciens
|
| 105 | c3h7aC_ |
|
not modelled |
88.9 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of short-chain dehydrogenase from2 rhodopseudomonas palustris
|
| 106 | d1o5ia_ |
|
not modelled |
88.9 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 107 | c3nywD_ |
|
not modelled |
88.8 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a betaketoacyl-[acp] reductase (fabg) from2 bacteroides thetaiotaomicron
|
| 108 | c1ofgF_ |
|
not modelled |
88.8 |
16 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
| 109 | c1h6dL_ |
|
not modelled |
88.8 |
16 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
| 110 | d2f1ka2 |
|
not modelled |
88.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 111 | c2ekqB_ |
|
not modelled |
88.6 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-deoxy-d-gluconate 3-dehydrogenase;
PDBTitle: structure of tt0495 protein from thermus thermophilus
|
| 112 | c2ehdB_ |
|
not modelled |
88.4 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, short-chain dehydrogenase/reductase family;
PDBTitle: crystal structure analysis of oxidoreductase
|
| 113 | c2vouA_ |
|
not modelled |
88.3 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,6-dihydroxypyridine hydroxylase;
PDBTitle: structure of 2,6-dihydroxypyridine-3-hydroxylase from2 arthrobacter nicotinovorans
|
| 114 | c2pk3B_ |
|
not modelled |
88.3 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-6-deoxy-d-lyxo-4-hexulose reductase;
PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
|
| 115 | d2iida1 |
|
not modelled |
88.2 |
15 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 116 | c2p68A_ |
|
not modelled |
88.2 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of aq_1716 from aquifex aeolicus vf5
|
| 117 | c3v2hB_ |
|
not modelled |
88.1 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:d-beta-hydroxybutyrate dehydrogenase;
PDBTitle: the crystal structure of d-beta-hydroxybutyrate dehydrogenase from2 sinorhizobium meliloti
|
| 118 | c2pd6D_ |
|
not modelled |
88.1 |
23 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:estradiol 17-beta-dehydrogenase 8;
PDBTitle: structure of human hydroxysteroid dehydrogenase type 8, hsd17b8
|
| 119 | d2a4ka1 |
|
not modelled |
88.1 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 120 | d1gega_ |
|
not modelled |
88.1 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |

