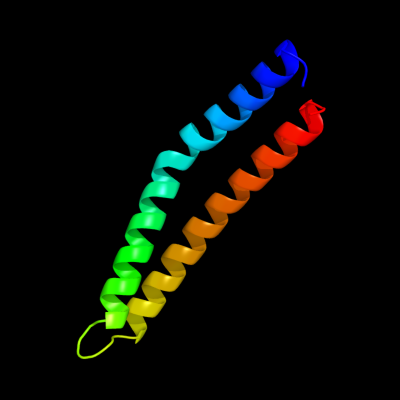
1 c3layF_
99.8
71
PDB header: metal binding proteinChain: F: PDB Molecule: zinc resistance-associated protein;PDBTitle: alpha-helical barrel formed by the decamer of the zinc resistance-2 associated protein (stm4172) from salmonella enterica subsp. enterica3 serovar typhimurium str. lt2
2 c3epvB_
99.3
16
PDB header: metal binding proteinChain: B: PDB Molecule: nickel and cobalt resistance protein cnrr;PDBTitle: x-ray structure of the metal-sensor cnrx in both the apo- and copper-2 bound forms
3 c3qzcA_
99.0
17
PDB header: signaling proteinChain: A: PDB Molecule: periplasmic protein cpxp;PDBTitle: structure of the periplasmic stress response protein cpxp
4 c3oeoD_
98.9
17
PDB header: signaling proteinChain: D: PDB Molecule: spheroplast protein y;PDBTitle: the crystal structure e. coli spy
5 c3o39A_
98.9
17
PDB header: chaperoneChain: A: PDB Molecule: periplasmic protein related to spheroblast formation;PDBTitle: crystal structure of spy
6 c3itfA_
98.9
14
PDB header: signaling proteinChain: A: PDB Molecule: periplasmic adaptor protein cpxp;PDBTitle: structural basis for the inhibitory function of the cpxp adaptor2 protein
7 d1gqea_
83.6
22
Fold: Release factorSuperfamily: Release factorFamily: Release factor8 c1j1eC_
78.7
17
PDB header: contractile proteinChain: C: PDB Molecule: troponin i;PDBTitle: crystal structure of the 52kda domain of human cardiac2 troponin in the ca2+ saturated form
9 c2w6aB_
77.0
20
PDB header: signaling proteinChain: B: PDB Molecule: arf gtpase-activating protein git1;PDBTitle: x-ray structure of the dimeric git1 coiled-coil domain
10 c2gl2B_
75.4
15
PDB header: cell adhesionChain: B: PDB Molecule: adhesion a;PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,2 y69g) of bacterial adhesin fada
11 c1ei3E_
73.7
5
PDB header: PDB COMPND: 12 c3ojaB_
72.3
15
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
13 c3ghgK_
70.9
8
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
14 c1deqF_
68.7
12
PDB header: PDB COMPND: 15 c3bt6B_
68.0
21
PDB header: viral proteinChain: B: PDB Molecule: influenza b hemagglutinin (ha);PDBTitle: crystal structure of influenza b virus hemagglutinin
16 c2ke4A_
67.8
16
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
17 c1deqO_
67.7
8
PDB header: PDB COMPND: 18 c3hnwB_
67.3
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a basic coiled-coil protein of unknown function2 from eubacterium eligens atcc 27750
19 d1cuna2
67.1
16
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat20 c1yv0I_
66.8
21
PDB header: contractile proteinChain: I: PDB Molecule: troponin i, fast skeletal muscle;PDBTitle: crystal structure of skeletal muscle troponin in the ca2+-2 free state
21 c2gd7B_
not modelled
63.5
12
PDB header: transcriptionChain: B: PDB Molecule: hexim1 protein;PDBTitle: the structure of the cyclin t-binding domain of hexim12 reveals the molecular basis for regulation of3 transcription elongation
22 d1s35a2
not modelled
63.4
16
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat23 c3kltB_
not modelled
59.4
13
PDB header: structural proteinChain: B: PDB Molecule: vimentin;PDBTitle: crystal structure of a vimentin fragment
24 c2w9yA_
not modelled
53.6
12
PDB header: lipid transportChain: A: PDB Molecule: fatty acid/retinol binding protein protein 7,PDBTitle: the structure of the lipid binding protein ce-far-7 from2 caenorhabditis elegans
25 c2iakA_
not modelled
53.3
16
PDB header: cell adhesionChain: A: PDB Molecule: bullous pemphigoid antigen 1, isoform 5;PDBTitle: crystal structure of a protease resistant fragment of the plakin2 domain of bullous pemphigoid antigen1 (bpag1)
26 d2spca_
not modelled
51.1
19
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat27 c2v7sA_
not modelled
50.2
19
PDB header: unknown functionChain: A: PDB Molecule: probable conserved lipoprotein lppa;PDBTitle: crystal structure of the putative lipoprotein lppa from2 mycobacterium tuberculosis
28 d2oeza1
not modelled
49.1
23
Fold: YacF-likeSuperfamily: YacF-likeFamily: YacF-like29 c2jo8B_
not modelled
49.0
10
PDB header: transferaseChain: B: PDB Molecule: serine/threonine-protein kinase 4;PDBTitle: solution structure of c-terminal domain of human mammalian2 sterile 20-like kinase 1 (mst1)
30 c3m9bK_
not modelled
48.1
17
PDB header: chaperoneChain: K: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
31 d1vqov1
not modelled
48.0
17
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)32 c2a3dA_
not modelled
46.5
23
PDB header: three-helix bundleChain: A: PDB Molecule: protein (de novo three-helix bundle);PDBTitle: solution structure of a de novo designed single chain three-2 helix bundle (a3d)
33 c3d5cX_
not modelled
46.5
17
PDB header: ribosomeChain: X: PDB Molecule: peptide chain release factor 1;PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 30s subunit, release factor 1 (rf1), two trna, and3 mrna molecules of the second 70s ribosome. the entire crystal4 structure contains two 70s ribosomes as described in remark 400.
34 c3ls1A_
not modelled
45.4
18
PDB header: photosynthesisChain: A: PDB Molecule: sll1638 protein;PDBTitle: crystal structure of cyanobacterial psbq from synechocystis2 sp. pcc 6803 complexed with zn2+
35 d1quua1
not modelled
44.3
16
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat36 c1wpaA_
not modelled
42.6
17
PDB header: cell adhesionChain: A: PDB Molecule: occludin;PDBTitle: 1.5 angstrom crystal structure of human occludin fragment2 413-522
37 c1xzqA_
not modelled
40.9
13
PDB header: hydrolaseChain: A: PDB Molecule: probable trna modification gtpase trme;PDBTitle: structure of the gtp-binding protein trme from thermotoga2 maritima complexed with 5-formyl-thf
38 c2xdjF_
not modelled
40.8
12
PDB header: unknown functionChain: F: PDB Molecule: uncharacterized protein ybgf;PDBTitle: crystal structure of the n-terminal domain of e.coli ybgf
39 c2js5B_
not modelled
40.1
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of protein q60c73_metca. northeast structural2 genomics consortium target mcr1
40 d1u5pa1
not modelled
39.7
16
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat41 c2fxmB_
not modelled
38.5
10
PDB header: contractile proteinChain: B: PDB Molecule: myosin heavy chain, cardiac muscle beta isoform;PDBTitle: structure of the human beta-myosin s2 fragment
42 c1deqD_
not modelled
38.3
17
PDB header: PDB COMPND: 43 c2j375_
not modelled
38.1
16
PDB header: ribosomeChain: 5: PDB Molecule: ribosomal protein l35;PDBTitle: model of mammalian srp bound to 80s rncs
44 c2eqbC_
not modelled
38.0
13
PDB header: endocytosis/exocytosisChain: C: PDB Molecule: rab guanine nucleotide exchange factor sec2;PDBTitle: crystal structure of the rab gtpase sec4p, the sec2p gef2 domain, and phosphate complex
45 d1u5pa2
not modelled
37.8
14
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat46 c3kbtA_
not modelled
37.4
13
PDB header: structural proteinChain: A: PDB Molecule: spectrin beta chain, erythrocyte;PDBTitle: crystal structure of the ankyrin binding domain of human erythroid2 beta spectrin (repeats 13-15) in complex with the spectrin binding3 domain of human erythroid ankyrin (zu5-ank)
47 d1s35a1
not modelled
36.4
9
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat48 d1cxzb_
not modelled
36.3
14
Fold: Long alpha-hairpinSuperfamily: HR1 repeatFamily: HR1 repeat49 c3hizB_
not modelled
36.2
15
PDB header: transferase/oncoproteinChain: B: PDB Molecule: phosphatidylinositol 3-kinase regulatory subunitPDBTitle: crystal structure of p110alpha h1047r mutant in complex with2 nish2 of p85alpha
50 c2oszA_
not modelled
35.8
10
PDB header: structural proteinChain: A: PDB Molecule: nucleoporin p58/p45;PDBTitle: structure of nup58/45 suggests flexible nuclear pore diameter by2 intermolecular sliding
51 c2c5iT_
not modelled
35.7
20
PDB header: protein transportChain: T: PDB Molecule: t-snare affecting a late golgi compartmentPDBTitle: n-terminal domain of tlg1 complexed with n-terminus of2 vps51 in distorted conformation
52 c2p2uA_
not modelled
35.4
15
PDB header: dna binding proteinChain: A: PDB Molecule: host-nuclease inhibitor protein gam, putative;PDBTitle: crystal structure of putative host-nuclease inhibitor2 protein gam from desulfovibrio vulgaris
53 c2zkrv_
not modelled
34.2
11
PDB header: ribosomal protein/rnaChain: V: PDB Molecule: rna expansion segment es9 part2;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
54 c3csxA_
not modelled
33.4
18
PDB header: metal binding protein,unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structural characterization of a protein in the duf6832 family- crystal structure of cce_0567 from the3 cyanobacterium cyanothece 51142.
55 d1st6a3
not modelled
32.3
19
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin56 c3ol1A_
not modelled
30.7
8
PDB header: structural proteinChain: A: PDB Molecule: vimentin;PDBTitle: crystal structure of vimentin (fragment 144-251) from homo sapiens,2 northeast structural genomics consortium target hr4796b
57 d1uklc_
not modelled
30.6
22
Fold: HLH-likeSuperfamily: HLH, helix-loop-helix DNA-binding domainFamily: HLH, helix-loop-helix DNA-binding domain58 d1olma1
not modelled
29.8
17
Fold: RuvA C-terminal domain-likeSuperfamily: CRAL/TRIO N-terminal domainFamily: CRAL/TRIO N-terminal domain59 c3f31B_
not modelled
29.4
17
PDB header: actin binding, structural proteinChain: B: PDB Molecule: spectrin alpha chain, brain;PDBTitle: crystal structure of the n-terminal region of alphaii-spectrin2 tetramerization domain
60 c3m0dC_
not modelled
29.4
13
PDB header: signaling proteinChain: C: PDB Molecule: tnf receptor-associated factor 1;PDBTitle: crystal structure of the traf1:traf2:ciap2 complex
61 c2ihr1_
not modelled
28.8
26
PDB header: translationChain: 1: PDB Molecule: peptide chain release factor 2;PDBTitle: rf2 of thermus thermophilus
62 c3gehA_
not modelled
28.7
16
PDB header: hydrolaseChain: A: PDB Molecule: trna modification gtpase mnme;PDBTitle: crystal structure of mnme from nostoc in complex with gdp, folinic2 acid and zn
63 c3hfdA_
not modelled
28.6
9
PDB header: chaperone, protein transportChain: A: PDB Molecule: nucleosome assembly protein 1;PDBTitle: nucleosome assembly protein 1 from plasmodium knowlesi
64 c2j8pA_
not modelled
27.9
22
PDB header: nuclear proteinChain: A: PDB Molecule: cleavage stimulation factor 64 kda subunit;PDBTitle: nmr structure of c-terminal domain of human cstf-64
65 c3fs3A_
not modelled
27.3
11
PDB header: chaperoneChain: A: PDB Molecule: nucleosome assembly protein 1, putative;PDBTitle: crystal structure of malaria parasite nucleosome assembly protein2 (nap)
66 c2an7A_
not modelled
26.9
15
PDB header: dna binding proteinChain: A: PDB Molecule: protein pard;PDBTitle: solution structure of the bacterial antidote pard
67 c3ni0A_
not modelled
26.9
20
PDB header: immune systemChain: A: PDB Molecule: bone marrow stromal antigen 2;PDBTitle: crystal structure of mouse bst-2/tetherin ectodomain
68 c1xawA_
not modelled
26.6
19
PDB header: cell adhesionChain: A: PDB Molecule: occludin;PDBTitle: crystal structure of the cytoplasmic distal c-terminal2 domain of occludin
69 c1s35A_
not modelled
26.4
16
PDB header: structural proteinChain: A: PDB Molecule: spectrin beta chain, erythrocyte;PDBTitle: crystal structure of repeats 8 and 9 of human erythroid2 spectrin
70 d1st6a4
not modelled
26.0
27
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin71 d1ydxa1
not modelled
25.6
13
Fold: DNA methylase specificity domainSuperfamily: DNA methylase specificity domainFamily: Type I restriction modification DNA specificity domain72 c3bhpA_
not modelled
25.5
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0291 protein ynzc;PDBTitle: crystal structure of upf0291 protein ynzc from bacillus2 subtilis at resolution 2.0 a. northeast structural3 genomics consortium target sr384
73 d1akha_
not modelled
25.4
7
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain74 c3ghgD_
not modelled
25.0
15
PDB header: blood clottingChain: D: PDB Molecule: fibrinogen alpha chain;PDBTitle: crystal structure of human fibrinogen
75 d1lvaa3
not modelled
24.7
5
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal fragment of elongation factor SelB76 c2no2A_
not modelled
24.5
14
PDB header: cell adhesionChain: A: PDB Molecule: huntingtin-interacting protein 1;PDBTitle: crystal structure of the dllrkn-containing coiled-coil2 domain of huntingtin-interacting protein 1
77 c3u59C_
not modelled
24.3
10
PDB header: contractile proteinChain: C: PDB Molecule: tropomyosin beta chain;PDBTitle: n-terminal 98-aa fragment of smooth muscle tropomyosin beta
78 c3qh9A_
not modelled
23.9
13
PDB header: structural proteinChain: A: PDB Molecule: liprin-beta-2;PDBTitle: human liprin-beta2 coiled-coil
79 d1ykhb1
not modelled
23.7
16
Fold: Mediator hinge subcomplex-likeSuperfamily: Mediator hinge subcomplex-likeFamily: CSE2-like80 d1hcia4
not modelled
23.5
12
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat81 c3m5gD_
not modelled
22.4
16
PDB header: viral proteinChain: D: PDB Molecule: hemagglutinin;PDBTitle: crystal structure of a h7 influenza virus hemagglutinin
82 c2nrjA_
not modelled
22.2
14
PDB header: toxinChain: A: PDB Molecule: hbl b protein;PDBTitle: crystal structure of hemolysin binding component from2 bacillus cereus
83 c3a7mA_
not modelled
21.9
12
PDB header: gene regulation, chaperoneChain: A: PDB Molecule: flagellar protein flit;PDBTitle: structure of flit, the flagellar type iii chaperone for flid
84 c2q9qC_
not modelled
21.9
16
PDB header: replicationChain: C: PDB Molecule: dna replication complex gins protein psf1;PDBTitle: the crystal structure of full length human gins complex
85 c2jeeA_
not modelled
21.8
14
PDB header: cell cycleChain: A: PDB Molecule: yiiu;PDBTitle: xray structure of e. coli yiiu
86 c2xusA_
not modelled
21.6
20
PDB header: protein bindingChain: A: PDB Molecule: breast cancer metastasis-suppressor 1;PDBTitle: crystal structure of the brms1 n-terminal region
87 c1jccC_
not modelled
21.5
11
PDB header: membrane proteinChain: C: PDB Molecule: major outer membrane lipoprotein;PDBTitle: crystal structure of a novel alanine-zipper trimer at 1.7 a2 resolution, v13a,l16a,v20a,l23a,v27a,m30a,v34a mutations
88 d2e9xa1
not modelled
21.5
16
Fold: GINS helical bundle-likeSuperfamily: GINS helical bundle-likeFamily: PSF1 N-terminal domain-like89 c2v6lI_
not modelled
21.3
20
PDB header: protein transportChain: I: PDB Molecule: mxih;PDBTitle: molecular model of a type iii secretion system needle
90 c1gk6B_
not modelled
21.3
10
PDB header: vimentinChain: B: PDB Molecule: vimentin;PDBTitle: human vimentin coil 2b fragment linked to gcn4 leucine2 zipper (z2b)
91 c1m1jA_
not modelled
21.2
17
PDB header: blood clottingChain: A: PDB Molecule: fibrinogen alpha subunit;PDBTitle: crystal structure of native chicken fibrinogen with two different2 bound ligands
92 c1avyA_
not modelled
20.0
30
PDB header: coiled coilChain: A: PDB Molecule: fibritin;PDBTitle: fibritin deletion mutant m (bacteriophage t4)
93 c3geiB_
not modelled
19.8
18
PDB header: hydrolaseChain: B: PDB Molecule: trna modification gtpase mnme;PDBTitle: crystal structure of mnme from chlorobium tepidum in complex2 with gcp
94 d2hgq11
not modelled
19.7
13
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)95 c1cunC_
not modelled
19.7
16
PDB header: structural proteinChain: C: PDB Molecule: protein (alpha spectrin);PDBTitle: crystal structure of repeats 16 and 17 of chicken brain2 alpha spectrin
96 c3b7qA_
not modelled
19.4
19
PDB header: signaling proteinChain: A: PDB Molecule: uncharacterized protein ykl091c;PDBTitle: crystal structure of yeast sec14 homolog sfh1 in complex with2 phosphatidylcholine
97 c1ox3A_
not modelled
19.2
26
PDB header: chaperoneChain: A: PDB Molecule: fibritin;PDBTitle: crystal structure of mini-fibritin
98 c1stzB_
not modelled
18.7
18
PDB header: transcriptionChain: B: PDB Molecule: heat-inducible transcription repressor hrca homolog;PDBTitle: crystal structure of a hypothetical protein at 2.2 a resolution
99 c1jsdB_
not modelled
18.6
18
PDB header: viral proteinChain: B: PDB Molecule: haemagglutinin (ha2 chain);PDBTitle: crystal structure of swine h9 haemagglutinin