1 c2jc7A_
96.6
14
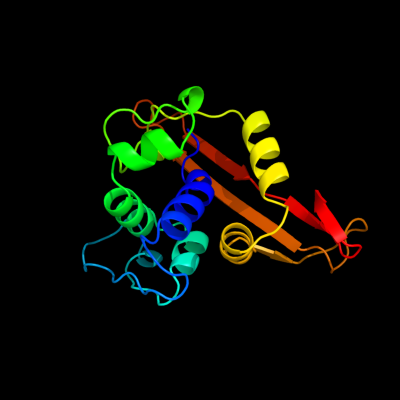
PDB header: hydrolaseChain: A: PDB Molecule: beta-lactamase oxa-24;PDBTitle: the crystal structure of the carbapenemase oxa-24 reveals2 new insights into the mechanism of carbapenem-hydrolysis
2 c3oc2A_
96.3
17
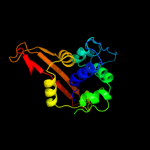
PDB header: penicillin-binding proteinChain: A: PDB Molecule: penicillin-binding protein 3;PDBTitle: crystal structure of penicillin-binding protein 3 from pseudomonas2 aeruginosa
3 c3if6C_
96.1
20
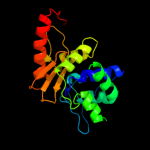
PDB header: hydrolaseChain: C: PDB Molecule: oxa-46 oxacillinase;PDBTitle: crystal structure of oxa-46 beta-lactamase from p.2 aeruginosa
4 c3lo7A_
96.0
23
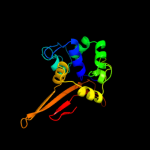
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding protein a;PDBTitle: crystal structure of pbpa from mycobacterium tuberculosis
5 d1k55a_
95.4
15
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase6 d1nrfa_
95.1
14
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase7 c3hbrD_
94.6
16
PDB header: hydrolaseChain: D: PDB Molecule: oxa-48;PDBTitle: crystal structure of oxa-48 beta-lactamase
8 d1xa1a_
94.5
16
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase9 c3equB_
94.3
17
PDB header: biosynthetic proteinChain: B: PDB Molecule: penicillin-binding protein 2;PDBTitle: crystal structure of penicillin-binding protein 2 from neisseria2 gonorrhoeae
10 c3udiA_
94.1
19
PDB header: penicillin-binding protein/antibioticChain: A: PDB Molecule: penicillin-binding protein 1a;PDBTitle: crystal structure of acinetobacter baumannii pbp1a in complex with2 penicillin g
11 d1k38a_
93.9
18
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase12 c2iwdA_
93.7
12
PDB header: antibiotic resistanceChain: A: PDB Molecule: methicillin resistance mecr1 protein;PDBTitle: oxacilloyl-acylated mecr1 extracellular antibiotic-sensor2 domain.
13 c3ue3A_
93.1
19
PDB header: transferaseChain: A: PDB Molecule: septum formation, penicillin binding protein 3,PDBTitle: crystal structure of acinetobacter baumanni pbp3
14 c3pbqA_
92.8
19
PDB header: hydrolase/antibioticChain: A: PDB Molecule: penicillin-binding protein 3;PDBTitle: crystal structure of pbp3 complexed with imipenem
15 d1vqqa3
91.9
16
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase16 c2olvA_
89.5
16
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding protein 2;PDBTitle: structural insight into the transglycosylation step of bacterial cell2 wall biosynthesis : donor ligand complex
17 d1rp5a4
88.8
16
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase18 d1pyya4
88.1
14
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase19 c1qmfA_
87.7
16
PDB header: peptidoglycan synthesisChain: A: PDB Molecule: penicillin-binding protein 2x;PDBTitle: penicillin-binding protein 2x (pbp-2x) acyl-enzyme complex
20 c1pmdA_
86.9
16
PDB header: peptidoglycan synthesisChain: A: PDB Molecule: peptidoglycan synthesis multifunctional enzyme;PDBTitle: penicillin-binding protein 2x (pbp-2x)
21 c2bg1A_
not modelled
86.6
17
PDB header: peptidoglycanChain: A: PDB Molecule: penicillin-binding protein 1b;PDBTitle: active site restructuring regulates ligand recognition in2 classa penicillin-binding proteins (pbps)
22 d1mr1c_
not modelled
84.0
30
Fold: SAND domain-likeSuperfamily: SAND domain-likeFamily: SMAD4-binding domain of oncoprotein Ski23 c3fwlA_
not modelled
82.9
16
PDB header: transferase, hydrolaseChain: A: PDB Molecule: penicillin-binding protein 1b;PDBTitle: crystal structure of the full-length transglycosylase pbp1b2 from escherichia coli
24 d2c5wb1
not modelled
82.6
18
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase25 c3dwkC_
not modelled
81.1
19
PDB header: transferaseChain: C: PDB Molecule: penicillin-binding protein 2;PDBTitle: identification of dynamic structural motifs involved in2 peptidoglycan glycosyltransfer
26 d2bg1a1
not modelled
79.7
18
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase27 c2wadB_
not modelled
78.0
21
PDB header: peptide binding proteinChain: B: PDB Molecule: penicillin-binding protein 2b;PDBTitle: penicillin-binding protein 2b (pbp-2b) from streptococcus2 pneumoniae (strain 5204)
28 c2jciA_
not modelled
77.9
19
PDB header: drug-binding proteinChain: A: PDB Molecule: penicillin-binding protein 1b;PDBTitle: structural insights into the catalytic mechanism and the2 role of streptococcus pneumoniae pbp1b
29 c1mwuA_
not modelled
70.8
16
PDB header: biosynthetic proteinChain: A: PDB Molecule: penicillin-binding protein 2a;PDBTitle: structure of methicillin acyl-penicillin binding protein 2a2 from methicillin resistant staphylococcus aureus strain3 27r at 2.60 a resolution.
30 c1t6sB_
63.6
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of a conserved hypothetical protein from chlorobium2 tepidum
31 c2z99A_
63.2
16
PDB header: cell cycleChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of scpb from mycobacterium tuberculosis
32 d2olua2
not modelled
61.1
18
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase33 c3dtkA_
52.0
31
PDB header: gene regulationChain: A: PDB Molecule: irre protein;PDBTitle: crystal structure of the irre protein, a central regulator2 of dna damage repair in deinococcaceae
34 c2xftA_
not modelled
41.4
17
PDB header: hydrolaseChain: A: PDB Molecule: orf12;PDBTitle: structural and mechanistic studies on a cephalosporin esterase from2 the clavulanic acid biosynthesis pathway
35 c3gwrA_
not modelled
41.1
8
PDB header: protein bindingChain: A: PDB Molecule: putative calcium/calmodulin-dependent protein kinase typePDBTitle: crystal structure of putative calcium/calmodulin-dependent protein2 kinase type ii association domain (yp_315894.1) from thiobacillus3 denitrificans atcc 25259 at 2.00 a resolution
36 d3elga1
not modelled
35.4
26
Fold: BLIP-likeSuperfamily: BT0923-likeFamily: BT0923-like37 d1m6ka_
not modelled
32.8
14
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase38 d1dy6a_
not modelled
31.4
19
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase39 c2hcbC_
not modelled
30.4
35
PDB header: replicationChain: C: PDB Molecule: chromosomal replication initiator protein dnaa;PDBTitle: structure of amppcp-bound dnaa from aquifex aeolicus
40 c2dafA_
not modelled
28.1
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: flj35834 protein;PDBTitle: solution structure of the novel identified ubiquitin-like2 domain in the human hypothetical protein flj35834
41 c3k7cC_
not modelled
27.8
25
PDB header: protein bindingChain: C: PDB Molecule: putative ntf2-like transpeptidase;PDBTitle: crystal structure of putative ntf2-like transpeptidase (np_281412.1)2 from campylobacter jejuni at 2.00 a resolution
42 d2pf5a1
not modelled
27.6
50
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Link domain43 c1aduB_
not modelled
25.7
28
PDB header: dna-binding proteinChain: B: PDB Molecule: adenovirus single-stranded dna-binding protein;PDBTitle: early e2a dna-binding protein
44 c1wg8B_
not modelled
25.6
40
PDB header: transferaseChain: B: PDB Molecule: predicted s-adenosylmethionine-dependentPDBTitle: crystal structure of a predicted s-adenosylmethionine-2 dependent methyltransferase tt1512 from thermus3 thermophilus hb8.
45 c1m6yA_
not modelled
23.3
33
PDB header: transferaseChain: A: PDB Molecule: s-adenosyl-methyltransferase mraw;PDBTitle: crystal structure analysis of tm0872, a putative sam-2 dependent methyltransferase, complexed with sah
46 d3duea1
not modelled
23.3
29
Fold: BLIP-likeSuperfamily: BT0923-likeFamily: BT0923-like47 d3cnxa1
not modelled
22.5
13
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: SAV4671-like48 c1zb7A_
not modelled
20.2
28
PDB header: toxinChain: A: PDB Molecule: neurotoxin;PDBTitle: crystal structure of botulinum neurotoxin type g light chain
49 d1l8qa2
not modelled
18.4
35
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain50 c3g5jA_
not modelled
18.3
18
PDB header: nucleotide binding proteinChain: A: PDB Molecule: putative atp/gtp binding protein;PDBTitle: crystal structure of n-terminal domain of putative atp/gtp binding2 protein from clostridium difficile 630
51 d1adta2
not modelled
17.2
24
Fold: Zn-binding domains of ADDBPSuperfamily: Zn-binding domains of ADDBPFamily: Zn-binding domains of ADDBP52 c3c0fB_
not modelled
14.3
32
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein af_1514;PDBTitle: crystal structure of a novel non-pfam protein af1514 from archeoglobus2 fulgidus dsm 4304 solved by s-sad using a cr x-ray source
53 d1k25a4
not modelled
13.7
23
Fold: beta-lactamase/transpeptidase-likeSuperfamily: beta-lactamase/transpeptidase-likeFamily: beta-Lactamase/D-ala carboxypeptidase54 c3u1nC_
not modelled
13.3
23
PDB header: hydrolaseChain: C: PDB Molecule: sam domain and hd domain-containing protein 1;PDBTitle: structure of the catalytic core of human samhd1
55 d1t3ka_
not modelled
12.2
33
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Cell cycle control phosphatase, catalytic domain56 c3fkaD_
not modelled
12.0
25
PDB header: unknown functionChain: D: PDB Molecule: uncharacterized ntf-2 like protein;PDBTitle: crystal structure of a ntf-2 like protein of unknown function2 (spo1084) from silicibacter pomeroyi dss-3 at 1.69 a resolution
57 c2c99A_
not modelled
11.7
21
PDB header: transcription regulationChain: A: PDB Molecule: psp operon transcriptional activator;PDBTitle: structural basis of the nucleotide driven conformational2 changes in the aaa domain of transcription activator pspf
58 d1kyfa2
not modelled
11.7
26
Fold: Subdomain of clathrin and coatomer appendage domainSuperfamily: Subdomain of clathrin and coatomer appendage domainFamily: Clathrin adaptor appendage, alpha and beta chain-specific domain59 d1jqna_
not modelled
11.7
21
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate carboxylase60 c1xnjB_
not modelled
11.4
24
PDB header: transferaseChain: B: PDB Molecule: bifunctional 3'-phosphoadenosine 5'-phosphosulfatePDBTitle: aps complex of human paps synthetase 1
61 d2aq2b1
not modelled
11.4
28
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Superantigen toxins, N-terminal domain62 d1mhyd_
not modelled
11.0
18
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like63 d1p5dx4
not modelled
10.9
18
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain64 c2jx8A_
not modelled
10.7
38
PDB header: transcriptionChain: A: PDB Molecule: phosphorylated ctd-interacting factor 1;PDBTitle: solution structure of hpcif1 ww domain
65 d1whqa_
not modelled
10.4
28
Fold: dsRBD-likeSuperfamily: dsRNA-binding domain-likeFamily: Double-stranded RNA-binding domain (dsRBD)66 c1jqoA_
not modelled
10.2
18
PDB header: lyaseChain: A: PDB Molecule: phosphoenolpyruvate carboxylase;PDBTitle: crystal structure of c4-form phosphoenolpyruvate carboxylase from2 maize
67 d1jqoa_
not modelled
10.2
18
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate carboxylase68 c2r44A_
not modelled
10.1
21
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative atpase (chu_0153) from cytophaga2 hutchinsonii atcc 33406 at 2.00 a resolution
69 c3nfiB_
not modelled
10.0
14
PDB header: dna binding protein, transcriptionChain: B: PDB Molecule: dna-directed rna polymerase i subunit rpa49;PDBTitle: crystal structure of tandem winged helix domain of rna polymerase i2 subunit a49
70 c2givA_
not modelled
10.0
26
PDB header: transferaseChain: A: PDB Molecule: probable histone acetyltransferase myst1;PDBTitle: human myst histone acetyltransferase 1
71 d2giva1
not modelled
10.0
26
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: N-acetyl transferase, NAT72 c3dukD_
not modelled
9.7
25
PDB header: unknown functionChain: D: PDB Molecule: ntf2-like protein of unknown function;PDBTitle: crystal structure of a ntf2-like protein of unknown function2 (mfla_0564) from methylobacillus flagellatus kt at 2.200 a resolution
73 d1qvra2
not modelled
9.7
17
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Extended AAA-ATPase domain74 c3hfkB_
not modelled
9.7
8
PDB header: isomeraseChain: B: PDB Molecule: 4-methylmuconolactone methylisomerase;PDBTitle: crystal structure of 4-methylmuconolactone methylisomerase2 (h52a) in complex with 4-methylmuconolactone
75 c3nhvE_
not modelled
9.6
21
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: bh2092 protein;PDBTitle: crystal structure of bh2092 protein from bacillus halodurans,2 northeast structural genomics consortium target bhr228f
76 c3iukB_
not modelled
9.3
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of putative bacterial protein of unknown function2 (duf885, pf05960.1, ) from arthrobacter aurescens tc1, reveals fold3 similar to that of m32 carboxypeptidases
77 c2jofA_
not modelled
9.1
60
PDB header: de novo proteinChain: A: PDB Molecule: trp-cage;PDBTitle: the trp-cage: optimizing the stability of a globular2 miniprotein
78 d1g8fa3
not modelled
9.1
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain79 c2q14A_
not modelled
9.0
23
PDB header: hydrolaseChain: A: PDB Molecule: phosphohydrolase;PDBTitle: crystal structure of phosphohydrolase (bt4208) from bacteroides2 thetaiotaomicron vpi-5482 at 2.20 a resolution
80 c3f7sA_
not modelled
8.7
5
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized ntf2-like protein;PDBTitle: crystal structure of a ntf2-like protein of unknown function (pp_4556)2 from pseudomonas putida kt2440 at 2.11 a resolution
81 d1fw8a_
not modelled
8.5
29
Fold: Phosphoglycerate kinaseSuperfamily: Phosphoglycerate kinaseFamily: Phosphoglycerate kinase82 d1k82a2
not modelled
8.4
17
Fold: N-terminal domain of MutM-like DNA repair proteinsSuperfamily: N-terminal domain of MutM-like DNA repair proteinsFamily: N-terminal domain of MutM-like DNA repair proteins83 c1wx4B_
not modelled
8.3
18
PDB header: oxidoreductase/metal transportChain: B: PDB Molecule: melc;PDBTitle: crystal structure of the oxy-form of the copper-bound streptomyces2 castaneoglobisporus tyrosinase complexed with a caddie protein3 prepared by the addition of dithiothreitol
84 c2g7rA_
not modelled
8.3
36
PDB header: hydrolaseChain: A: PDB Molecule: mucosa-associated lymphoid tissue lymphoma translocationPDBTitle: x-ray structure of the death domain of the human mucosa associated2 lymphoid tissue lymphoma translocation protein 1
85 c2ee7A_
not modelled
8.2
20
PDB header: structural proteinChain: A: PDB Molecule: sperm flagellar protein 1;PDBTitle: solution structure of the ch domain from human sperm2 flagellar protein 1
86 d1fnua1
not modelled
8.1
22
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Superantigen toxins, N-terminal domain87 c1c4gB_
not modelled
8.0
32
PDB header: transferaseChain: B: PDB Molecule: protein (alpha-d-glucose 1-phosphatePDBTitle: phosphoglucomutase vanadate based transition state analog2 complex
88 c2wuqA_
not modelled
7.8
16
PDB header: transcriptionChain: A: PDB Molecule: beta-lactamase regulatory protein blab;PDBTitle: crystal structure of blab protein from streptomyces cacaoi
89 d1yt8a4
not modelled
7.8
16
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)90 d2bgca1
not modelled
7.7
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like91 c2bgcA_
not modelled
7.6
16
PDB header: transcriptionChain: A: PDB Molecule: prfa;PDBTitle: prfa-g145s, a constitutive active mutant of the2 transcriptional regulator in l.monocytogenes
92 d2cb5a_
not modelled
7.5
27
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Papain-like93 d1yt8a1
not modelled
7.5
36
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Multidomain sulfurtransferase (rhodanese)94 d1b24a2
not modelled
7.5
26
Fold: Homing endonuclease-likeSuperfamily: Homing endonucleasesFamily: Group I mobile intron endonuclease95 d1e32a3
not modelled
7.3
19
Fold: Cdc48 domain 2-likeSuperfamily: Cdc48 domain 2-likeFamily: Cdc48 domain 2-like96 d2ux0a1
not modelled
7.3
12
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Association domain of calcium/calmodulin-dependent protein kinase type II alpha subunit, CAMK2A97 d2f86b1
not modelled
7.3
15
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Association domain of calcium/calmodulin-dependent protein kinase type II alpha subunit, CAMK2A98 c3nbxX_
not modelled
7.2
18
PDB header: hydrolaseChain: X: PDB Molecule: atpase rava;PDBTitle: crystal structure of e. coli rava (regulatory atpase variant a) in2 complex with adp
99 c3emeA_
not modelled
7.1
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: rhodanese-like domain protein;PDBTitle: crystal structure of rhodanese-like domain protein from2 staphylococcus aureus