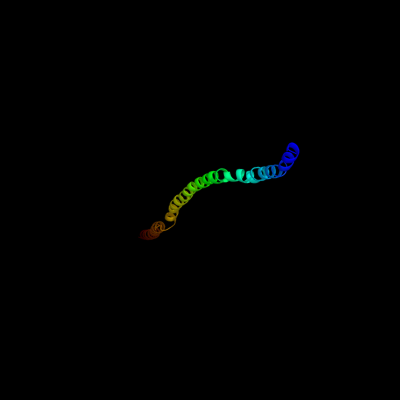
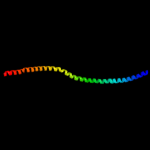
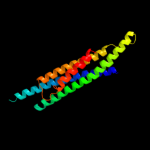
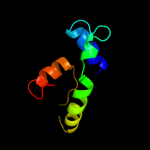
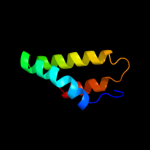
1 c3ghgK_
88.0
11
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
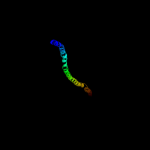
2 c1ei3E_
85.0
6
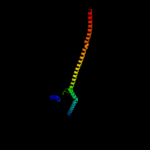
PDB header: PDB COMPND: 3 c3ojaB_
83.3
8
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
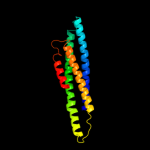
4 c1deqO_
82.7
14
PDB header: PDB COMPND: 5 c1deqF_
80.7
6
PDB header: PDB COMPND: 6 c3cwgA_
74.9
6
PDB header: transcriptionChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: unphosphorylated mouse stat3 core fragment
7 c1ei3C_
72.1
6
PDB header: PDB COMPND: 8 c1bf5A_
59.3
4
PDB header: gene regulation/dnaChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: tyrosine phosphorylated stat-1/dna complex
9 c2kr6A_
49.2
10
PDB header: hydrolaseChain: A: PDB Molecule: presenilin-1;PDBTitle: solution structure of presenilin-1 ctf subunit
10 c2a5yA_
47.1
12
PDB header: apoptosisChain: A: PDB Molecule: apoptosis regulator ced-9;PDBTitle: structure of a ced-4/ced-9 complex
11 c1bg1A_
38.5
5
PDB header: transcription/dnaChain: A: PDB Molecule: protein (transcription factor stat3b);PDBTitle: transcription factor stat3b/dna complex
12 c1f5nA_
27.9
9
PDB header: signaling proteinChain: A: PDB Molecule: interferon-induced guanylate-binding protein 1;PDBTitle: human guanylate binding protein-1 in complex with the gtp2 analogue, gmppnp.
13 c1ciiA_
27.5
8
PDB header: transmembrane proteinChain: A: PDB Molecule: colicin ia;PDBTitle: colicin ia
14 c2k8jX_
27.0
31
PDB header: viral proteinChain: X: PDB Molecule: p7tm2;PDBTitle: solution structure of hcv p7 tm2
15 d1ji6a3
23.6
16
Fold: Toxins' membrane translocation domainsSuperfamily: delta-Endotoxin (insectocide), N-terminal domainFamily: delta-Endotoxin (insectocide), N-terminal domain16 c1b9uA_
19.7
24
PDB header: hydrolaseChain: A: PDB Molecule: protein (atp synthase);PDBTitle: membrane domain of the subunit b of the e.coli atp synthase
17 d1r3jc_
17.4
6
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels18 c1mhsA_
16.8
10
PDB header: membrane protein, proton transportChain: A: PDB Molecule: plasma membrane atpase;PDBTitle: model of neurospora crassa proton atpase
19 d1o0la_
16.5
9
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death20 c3dtpA_
15.2
6
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain chimera of smooth andPDBTitle: tarantula heavy meromyosin obtained by flexible docking to2 tarantula muscle thick filament cryo-em 3d-map
21 c1yvlB_
not modelled
14.4
6
PDB header: signaling proteinChain: B: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: structure of unphosphorylated stat1
22 d2r6gf1
not modelled
13.8
15
Fold: MalF N-terminal region-likeSuperfamily: MalF N-terminal region-likeFamily: MalF N-terminal region-like23 c1g8xB_
not modelled
13.7
6
PDB header: structural proteinChain: B: PDB Molecule: myosin ii heavy chain fused to alpha-actinin 3;PDBTitle: structure of a genetically engineered molecular motor
24 c2xa0A_
not modelled
13.7
11
PDB header: apoptosisChain: A: PDB Molecule: apoptosis regulator bcl-2;PDBTitle: crystal structure of bcl-2 in complex with a bax bh32 peptide
25 c2eceA_
not modelled
13.4
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 462aa long hypothetical selenium-binding protein;PDBTitle: x-ray structure of hypothetical selenium-binding protein2 from sulfolobus tokodaii, st0059
26 c2wpqA_
not modelled
12.8
7
PDB header: membrane proteinChain: A: PDB Molecule: trimeric autotransporter adhesin fragment;PDBTitle: salmonella enterica sada 479-519 fused to gcn4 adaptors (2 sadak3, in-register fusion)
27 c2ki9A_
not modelled
12.7
30
PDB header: membrane proteinChain: A: PDB Molecule: cannabinoid receptor 2;PDBTitle: human cannabinoid receptor-2 helix 6
28 d2i5nl1
not modelled
12.0
11
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits29 c3ifxB_
not modelled
10.9
9
PDB header: membrane proteinChain: B: PDB Molecule: voltage-gated potassium channel;PDBTitle: crystal structure of the spin-labeled kcsa mutant v48r1
30 c2yv6A_
not modelled
10.7
20
PDB header: apoptosisChain: A: PDB Molecule: bcl-2 homologous antagonist/killer;PDBTitle: crystal structure of human bcl-2 family protein bak
31 c1ma1E_
not modelled
10.4
19
PDB header: oxidoreductaseChain: E: PDB Molecule: superoxide dismutase;PDBTitle: structure and properties of the atypical iron superoxide2 dismutase from methanobacterium thermoautotrophicum
32 c2rddB_
not modelled
10.4
30
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
33 c2nybC_
not modelled
9.8
10
PDB header: oxidoreductaseChain: C: PDB Molecule: superoxide dismutase [fe];PDBTitle: crystal structure of e.coli iron superoxide dismutase q69e2 at 1.1 angstrom resolution
34 d1rh1a2
not modelled
9.7
16
Fold: Toxins' membrane translocation domainsSuperfamily: ColicinFamily: Colicin35 d1vbga3
not modelled
9.7
6
Fold: ATP-graspSuperfamily: Glutathione synthetase ATP-binding domain-likeFamily: Pyruvate phosphate dikinase, N-terminal domain36 c1y67D_
not modelled
9.5
10
PDB header: oxidoreductaseChain: D: PDB Molecule: manganese superoxide dismutase;PDBTitle: crystal structure of manganese superoxide dismutase from2 deinococcus radiodurans
37 c1kkcB_
not modelled
9.4
10
PDB header: oxidoreductaseChain: B: PDB Molecule: manganese superoxide dismutase;PDBTitle: crystal structure of aspergillus fumigatus mnsod
38 d1y9ia_
not modelled
9.3
9
Fold: YutG-likeSuperfamily: YutG-likeFamily: YutG-like39 c3k07A_
not modelled
9.3
8
PDB header: transport proteinChain: A: PDB Molecule: cation efflux system protein cusa;PDBTitle: crystal structure of cusa
40 c1jchC_
not modelled
9.2
8
PDB header: ribosome inhibitor, hydrolaseChain: C: PDB Molecule: colicin e3;PDBTitle: crystal structure of colicin e3 in complex with its immunity protein
41 c2o2fA_
not modelled
9.2
11
PDB header: apoptosisChain: A: PDB Molecule: apoptosis regulator bcl-2;PDBTitle: solution structure of the anti-apoptotic protein bcl-2 in2 complex with an acyl-sulfonamide-based ligand
42 d1uerc2
not modelled
9.1
14
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain43 c1avmA_
not modelled
9.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: superoxide dismutase;PDBTitle: the cambialistic superoxide dismutase (fe-sod) of p. shermanii2 coordinated by azide
44 d1wb8a2
not modelled
8.9
14
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain45 d1bm8a_
not modelled
8.9
8
Fold: DNA-binding domain of Mlu1-box binding protein MBP1Superfamily: DNA-binding domain of Mlu1-box binding protein MBP1Family: DNA-binding domain of Mlu1-box binding protein MBP146 c3js4C_
not modelled
8.8
14
PDB header: oxidoreductaseChain: C: PDB Molecule: superoxide dismutase;PDBTitle: crystal structure of iron superoxide dismutase from anaplasma2 phagocytophilum
47 d1zy3a1
not modelled
8.8
9
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death48 c2cw2B_
not modelled
8.7
19
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase 1;PDBTitle: crystal structure of superoxide dismutase from p. marinus
49 c1qnnD_
not modelled
8.6
14
PDB header: oxidoreductaseChain: D: PDB Molecule: superoxide dismutase;PDBTitle: cambialistic superoxide dismutase from porphyromonas2 gingivalis
50 c3rg9A_
not modelled
8.5
33
PDB header: oxidoreductase/oxidoreductase inhibitorChain: A: PDB Molecule: bifunctional dihydrofolate reductase-thymidylate synthase;PDBTitle: trypanosoma brucei dihydrofolate reductase (tbdhfr) in complex with2 wr99210
51 d2q7ra1
not modelled
8.4
10
Fold: MAPEG domain-likeSuperfamily: MAPEG domain-likeFamily: MAPEG domain52 c1n0nB_
not modelled
8.2
14
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase [mn];PDBTitle: catalytic and structural effects of amino-acid substitution at his302 in human manganese superoxide dismutase
53 c3lj9A_
not modelled
8.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: iron superoxide dismutase;PDBTitle: x-ray structure of the iron superoxide dismutase from2 pseudoalteromonas haloplanktis in complex with sodium azide
54 c2gpcB_
not modelled
8.1
10
PDB header: oxidoreductaseChain: B: PDB Molecule: iron superoxide dismutase;PDBTitle: the crystal structure of the enzyme fe-superoxide dismutase2 from trypanosoma cruzi
55 c3l9oA_
not modelled
8.0
8
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent rna helicase dob1;PDBTitle: crystal structure of mtr4, a co-factor of the nuclear exosome
56 c3ceiA_
not modelled
8.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: superoxide dismutase;PDBTitle: crystal structure of superoxide dismutase from helicobacter2 pylori
57 c3kdpG_
not modelled
7.9
12
PDB header: hydrolaseChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
58 c3kdpH_
not modelled
7.9
12
PDB header: hydrolaseChain: H: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
59 c3tqjB_
not modelled
7.9
14
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase [fe];PDBTitle: structure of the superoxide dismutase (fe) (sodb) from coxiella2 burnetii
60 c2a03A_
not modelled
7.8
5
PDB header: oxidoreductaseChain: A: PDB Molecule: fe-superoxide dismutase homolog;PDBTitle: superoxide dismutase protein from plasmodium berghei
61 c3nnqA_
not modelled
7.8
3
PDB header: viral proteinChain: A: PDB Molecule: n-terminal domain of moloney murine leukemia virusPDBTitle: crystal structure of the n-terminal domain of moloney murine leukemia2 virus integrase, northeast structural genomics consortium target or3
62 c1p7gL_
not modelled
7.7
14
PDB header: oxidoreductaseChain: L: PDB Molecule: superoxide dismutase;PDBTitle: crystal structure of superoxide dismutase from pyrobaculum2 aerophilum
63 d1ma1a2
not modelled
7.6
19
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain64 c1gv3B_
not modelled
7.6
14
PDB header: manganese superoxide dismutaseChain: B: PDB Molecule: manganese superoxide dismutase;PDBTitle: the 2.0 angstrom resolution structure of the catalytic2 portion of a cyanobacterial membrane-bound manganese3 superoxide dismutase
65 d1y67a2
not modelled
7.5
10
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain66 d1b06a2
not modelled
7.5
10
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain67 c2rcvA_
not modelled
7.5
19
PDB header: oxidoreductaseChain: A: PDB Molecule: superoxide dismutase [mn];PDBTitle: crystal structure of the bacillus subtilis superoxide2 dismutase
68 c2kncB_
not modelled
7.5
26
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
69 c2y6xA_
not modelled
7.5
13
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii 11 kd protein;PDBTitle: structure of psb27 from thermosynechococcus elongatus
70 c1unfX_
not modelled
7.5
24
PDB header: oxidoreductaseChain: X: PDB Molecule: iron superoxide dismutase;PDBTitle: the crystal structure of the eukaryotic fesod from vigna2 unguiculata suggests a new enzymatic mechanism
71 d1bxla_
not modelled
7.4
9
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death72 c3prqT_
not modelled
7.3
18
PDB header: photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 1 of 2). this file contains first monomer of psii3 dimer
73 c3bz1T_
not modelled
7.3
18
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 12 of 2). this file contains first monomer of psii dimer
74 c3bz2T_
not modelled
7.3
18
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of cyanobacterial photosystem ii (part 22 of 2). this file contains second monomer of psii dimer
75 c3prrT_
not modelled
7.3
18
PDB header: photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of cyanobacterial photosystem ii in complex with2 terbutryn (part 2 of 2). this file contains second monomer of psii3 dimer
76 c3a0bT_
not modelled
7.3
15
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of br-substituted photosystem ii complex
77 c3arct_
not modelled
7.3
15
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
78 c1dt0A_
not modelled
7.3
5
PDB header: oxidoreductaseChain: A: PDB Molecule: superoxide dismutase;PDBTitle: cloning, sequence, and crystallographic structure of2 recombinant iron superoxide dismutase from pseudomonas3 ovalis
79 c1en4C_
not modelled
7.3
14
PDB header: oxidoreductaseChain: C: PDB Molecule: manganese superoxide dismutase;PDBTitle: crystal structure analysis of the e. coli manganese2 superoxide dismutase q146h mutant
80 c1gn4B_
not modelled
7.3
19
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase;PDBTitle: h145e mutant of mycobacterium tuberculosis iron-superoxide2 dismutase.
81 c1s5lT_
not modelled
7.2
18
PDB header: photosynthesisChain: T: PDB Molecule: photosystem ii psbt protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
82 c1s5lt_
not modelled
7.2
18
PDB header: photosynthesisChain: T: PDB Molecule: photosystem ii psbt protein;PDBTitle: architecture of the photosynthetic oxygen evolving center
83 c2cw3A_
not modelled
7.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: iron superoxide dismutase;PDBTitle: x-ray structure of pmsod2, superoxide dismutase from2 perkinsus marinus
84 c1sseB_
not modelled
7.2
11
PDB header: transcription activatorChain: B: PDB Molecule: ap-1 like transcription factor yap1;PDBTitle: solution structure of the oxidized form of the yap1 redox2 domain
85 d1uera2
not modelled
7.2
14
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain86 d2axtt1
not modelled
7.2
18
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein T, PsbTFamily: PsbT-like87 c3a0bt_
not modelled
7.2
18
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of br-substituted photosystem ii complex
88 c2axtT_
not modelled
7.2
18
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center t protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
89 c2axtt_
not modelled
7.2
18
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center t protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
90 c3arcT_
not modelled
7.2
18
PDB header: electron transport, photosynthesisChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
91 c3a0ht_
not modelled
7.2
18
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of i-substituted photosystem ii complex
92 c3kziT_
not modelled
7.2
18
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
93 c3a0hT_
not modelled
7.2
18
PDB header: electron transportChain: T: PDB Molecule: photosystem ii reaction center protein t;PDBTitle: crystal structure of i-substituted photosystem ii complex
94 d1p7ga2
not modelled
7.1
14
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain95 d1juva_
not modelled
7.1
40
Fold: Dihydrofolate reductase-likeSuperfamily: Dihydrofolate reductase-likeFamily: Dihydrofolate reductases96 d1pq1a_
not modelled
7.0
9
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death97 c2i88A_
not modelled
7.0
14
PDB header: membrane proteinChain: A: PDB Molecule: colicin-e1;PDBTitle: crystal structure of the channel-forming domain of colicin2 e1
98 d1kkca2
not modelled
6.9
10
Fold: Fe,Mn superoxide dismutase (SOD), C-terminal domainSuperfamily: Fe,Mn superoxide dismutase (SOD), C-terminal domainFamily: Fe,Mn superoxide dismutase (SOD), C-terminal domain99 c3h1sB_
not modelled
6.9
19
PDB header: oxidoreductaseChain: B: PDB Molecule: superoxide dismutase;PDBTitle: crystal structure of superoxide dismutase from francisella tularensis2 subsp. tularensis schu s4