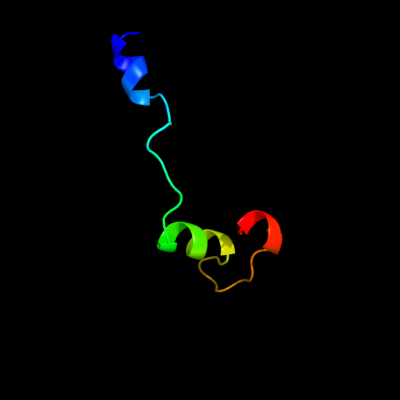
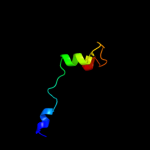
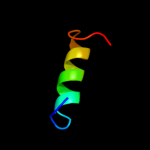
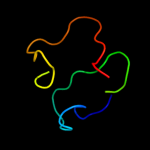
1 d1vlpa1
61.7
26
Fold: alpha/beta-HammerheadSuperfamily: Nicotinate/Quinolinate PRTase N-terminal domain-likeFamily: Monomeric nicotinate phosphoribosyltransferase N-terminal domain-like2 d1us7b_
45.1
32
Fold: Hsp90 co-chaperone CDC37Superfamily: Hsp90 co-chaperone CDC37Family: Hsp90 co-chaperone CDC373 c1us7B_
45.1
32
PDB header: chaperoneChain: B: PDB Molecule: hsp90 co-chaperone cdc37;PDBTitle: complex of hsp90 and p50
4 c2puyA_
39.5
26
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 21a;PDBTitle: crystal structure of the bhc80 phd finger
5 d2f7fa2
35.7
25
Fold: alpha/beta-HammerheadSuperfamily: Nicotinate/Quinolinate PRTase N-terminal domain-likeFamily: NadC N-terminal domain-like6 d1k0ga_
33.6
17
Fold: ADC synthaseSuperfamily: ADC synthaseFamily: ADC synthase7 d1ybea2
33.2
26
Fold: alpha/beta-HammerheadSuperfamily: Nicotinate/Quinolinate PRTase N-terminal domain-likeFamily: Monomeric nicotinate phosphoribosyltransferase N-terminal domain-like8 d1i7qa_
32.7
17
Fold: ADC synthaseSuperfamily: ADC synthaseFamily: ADC synthase9 d1i1qa_
31.5
16
Fold: ADC synthaseSuperfamily: ADC synthaseFamily: ADC synthase10 c1vlpA_
30.0
26
PDB header: transferaseChain: A: PDB Molecule: nicotinate phosphoribosyltransferase;PDBTitle: crystal structure of a putative nicotinate phosphoribosyltransferase2 (yor209c, npt1) from saccharomyces cerevisiae at 1.75 a resolution
11 d1j96a_
29.9
26
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)12 d1weqa_
28.6
24
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: PHD domain13 d1q5ma_
28.3
33
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)14 d1zbdb_
26.9
26
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: FYVE, a phosphatidylinositol-3-phosphate binding domain15 d1yira2
26.6
27
Fold: alpha/beta-HammerheadSuperfamily: Nicotinate/Quinolinate PRTase N-terminal domain-likeFamily: Monomeric nicotinate phosphoribosyltransferase N-terminal domain-like16 d1ah4a_
25.2
33
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)17 c3o37D_
24.0
38
PDB header: transcription/protein bindingChain: D: PDB Molecule: transcription intermediary factor 1-alpha;PDBTitle: crystal structure of trim24 phd-bromo complexed with h3(1-10)k42 peptide
18 d2alra_
22.6
29
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)19 d1hqta_
21.8
32
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)20 d3ctaa2
21.5
22
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin kinase-likeFamily: CTP-dependent riboflavin kinase-like21 c2riqA_
not modelled
20.6
28
PDB header: transferaseChain: A: PDB Molecule: poly [adp-ribose] polymerase 1;PDBTitle: crystal structure of the third zinc-binding domain of human parp-1
22 c3b4sA_
not modelled
20.0
56
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein luxt;PDBTitle: crystal structure of a luxt domain from vibrio2 parahaemolyticus rimd 2210633
23 d1mn4a_
not modelled
19.6
45
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: DNA-binding domain from NDT8024 c1yj7A_
not modelled
19.5
19
PDB header: protein transportChain: A: PDB Molecule: escj;PDBTitle: crystal structure of enteropathogenic e.coli (epec) type iii secretion2 system protein escj
25 c2obnA_
not modelled
19.2
26
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a duf1611 family protein (ava_3511) from anabaena2 variabilis atcc 29413 at 2.30 a resolution
26 c1xwhA_
not modelled
19.2
41
PDB header: transcriptionChain: A: PDB Molecule: autoimmune regulator;PDBTitle: nmr structure of the first phd finger of autoimmune2 regulator protein (aire1): insights into apeced
27 c2jvnA_
not modelled
18.7
30
PDB header: transferaseChain: A: PDB Molecule: poly [adp-ribose] polymerase 1;PDBTitle: domain c of human parp-1
28 d2g5fa1
not modelled
16.8
10
Fold: ADC synthaseSuperfamily: ADC synthaseFamily: ADC synthase29 d1q4qa_
not modelled
15.6
21
Fold: Inhibitor of apoptosis (IAP) repeatSuperfamily: Inhibitor of apoptosis (IAP) repeatFamily: Inhibitor of apoptosis (IAP) repeat30 d1iega_
not modelled
15.2
39
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin31 c3mhsE_
not modelled
15.0
33
PDB header: hydrolase/transcription regulator/proteiChain: E: PDB Molecule: saga-associated factor 73;PDBTitle: structure of the saga ubp8/sgf11/sus1/sgf73 dub module bound to2 ubiquitin aldehyde
32 c3h9mA_
not modelled
14.8
21
PDB header: lyaseChain: A: PDB Molecule: p-aminobenzoate synthetase, component i;PDBTitle: crystal structure of para-aminobenzoate synthetase,2 component i from cytophaga hutchinsonii
33 d2p6ra2
not modelled
14.6
26
Fold: Sec63 N-terminal domain-likeSuperfamily: Sec63 N-terminal domain-likeFamily: Achaeal helicase C-terminal domain34 d1afsa_
not modelled
14.4
28
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)35 c2im5C_
not modelled
14.3
26
PDB header: transferaseChain: C: PDB Molecule: nicotinate phosphoribosyltransferase;PDBTitle: crystal structure of nicotinate phosphoribosyltransferase2 from porphyromonas gingivalis
36 d1at3a_
not modelled
14.0
19
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin37 c2f6nA_
not modelled
13.7
21
PDB header: transcriptionChain: A: PDB Molecule: bromodomain phd finger transcription factor;PDBTitle: crystal structure of phd finger-linker-bromodomain fragment2 of human bptf in the free form
38 d2g0ta1
not modelled
13.7
24
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like39 d1us0a_
not modelled
13.3
38
Fold: TIM beta/alpha-barrelSuperfamily: NAD(P)-linked oxidoreductaseFamily: Aldo-keto reductases (NADP)40 c2i6yA_
not modelled
13.3
10
PDB header: lyaseChain: A: PDB Molecule: anthranilate synthase component i, putative;PDBTitle: structure and mechanism of mycobacterium tuberculosis salicylate2 synthase, mbti
41 c2fqxA_
not modelled
13.3
22
PDB header: transport proteinChain: A: PDB Molecule: membrane lipoprotein tmpc;PDBTitle: pnra from treponema pallidum complexed with guanosine
42 c2f7fA_
not modelled
13.3
25
PDB header: transferaseChain: A: PDB Molecule: nicotinate phosphoribosyltransferase, putative;PDBTitle: crystal structure of enterococcus faecalis putative nicotinate2 phosphoribosyltransferase, new york structural genomics consortium
43 d2pbka1
not modelled
13.1
32
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin44 d1ubdc2
not modelled
13.0
38
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H245 c2l43A_
not modelled
12.7
19
PDB header: transcriptionChain: A: PDB Molecule: n-teminal domain from histone h3.3, linker, phd1 domainPDBTitle: structural basis for histone code recognition by brpf2-phd1 finger
46 c2bs5A_
not modelled
12.5
47
PDB header: sugar binding proteinChain: A: PDB Molecule: fucose-binding lectin protein;PDBTitle: lectin from ralstonia solanacearum complexed with 2-2 fucosyllactose
47 d3efxd1
not modelled
12.4
43
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits48 d1vzva_
not modelled
12.4
21
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin49 d1vqow1
not modelled
12.4
13
Fold: Ribosomal protein L30p/L7eSuperfamily: Ribosomal protein L30p/L7eFamily: Ribosomal protein L30p/L7e50 c3bu8B_
not modelled
11.8
16
PDB header: dna binding proteinChain: B: PDB Molecule: telomeric repeat-binding factor 2;PDBTitle: crystal structure of trf2 trfh domain and tin2 peptide2 complex
51 d1qdla_
not modelled
11.6
18
Fold: ADC synthaseSuperfamily: ADC synthaseFamily: ADC synthase52 d1x3ha2
not modelled
11.3
36
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain53 c3pstA_
not modelled
11.1
17
PDB header: nuclear proteinChain: A: PDB Molecule: protein doa1;PDBTitle: crystal structure of pul and pfu(mutate) domain
54 d1xg8a_
not modelled
11.0
83
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: YuzD-like55 d2olua1
not modelled
10.9
10
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: PBP transglycosylase domain-like56 c3d6zA_
not modelled
10.6
33
PDB header: transcription regulator/dnaChain: A: PDB Molecule: multidrug-efflux transporter 1 regulator;PDBTitle: crystal structure of r275e mutant of bmrr bound to dna and rhodamine
57 c2euzA_
not modelled
10.6
50
PDB header: cell cycle/dnaChain: A: PDB Molecule: ndt80 protein;PDBTitle: structure of a ndt80-dna complex (mse mutant mc5t)
58 d1mnna_
not modelled
10.6
50
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: DNA-binding domain from NDT8059 c3elfA_
not modelled
10.5
20
PDB header: lyaseChain: A: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: structural characterization of tetrameric mycobacterium tuberculosis2 fructose 1,6-bisphosphate aldolase - substrate binding and catalysis3 mechanism of a class iia bacterial aldolase
60 d1o6ea_
not modelled
10.0
29
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin61 d3chbd_
not modelled
10.0
44
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits62 c3ln3A_
not modelled
10.0
39
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodiol dehydrogenase;PDBTitle: crystal structure of putative reductase (np_038806.2) from2 mus musculus at 1.18 a resolution
63 c2krcA_
not modelled
9.6
43
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase subunit delta;PDBTitle: solution structure of the n-terminal domain of bacillus2 subtilis delta subunit of rna polymerase
64 c2ysmA_
not modelled
9.5
23
PDB header: transferaseChain: A: PDB Molecule: myeloid/lymphoid or mixed-lineage leukemiaPDBTitle: solution structure of the first and second phd domain from2 myeloid/lymphoid or mixed-lineage leukemia protein 33 homolog
65 c2w0tA_
not modelled
9.4
31
PDB header: transcriptionChain: A: PDB Molecule: lethal(3)malignant brain tumor-like 2 protein;PDBTitle: solution structure of the fcs zinc finger domain of human2 lmbl2
66 c2l7pA_
not modelled
9.3
13
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase ashh2;PDBTitle: ashh2 a cw domain
67 c2a20A_
not modelled
9.3
26
PDB header: metal binding proteinChain: A: PDB Molecule: regulating synaptic membrane exocytosis proteinPDBTitle: solution structure of rim2 zinc finger domain
68 c2e6rA_
not modelled
9.3
24
PDB header: dna binding proteinChain: A: PDB Molecule: jumonji/arid domain-containing protein 1d;PDBTitle: solution structure of the phd domain in smcy protein
69 d1npba_
not modelled
9.0
31
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Antibiotic resistance proteins70 c3os6A_
not modelled
9.0
17
PDB header: isomeraseChain: A: PDB Molecule: isochorismate synthase dhbc;PDBTitle: crystal structure of putative 2,3-dihydroxybenzoate-specific2 isochorismate synthase, dhbc from bacillus anthracis.
71 d1olma1
not modelled
8.7
20
Fold: RuvA C-terminal domain-likeSuperfamily: CRAL/TRIO N-terminal domainFamily: CRAL/TRIO N-terminal domain72 d2af7a1
not modelled
8.6
16
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like73 d1kjna_
not modelled
8.4
29
Fold: Hypothetical protein MTH777 (MT0777)Superfamily: Hypothetical protein MTH777 (MT0777)Family: Hypothetical protein MTH777 (MT0777)74 d2h1ta1
not modelled
8.3
27
Fold: Spiral beta-rollSuperfamily: PA1994-likeFamily: PA1994-like75 d1c8aa2
not modelled
8.2
35
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain76 d1l0nk_
not modelled
8.2
33
Fold: Single transmembrane helixSuperfamily: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)77 d1i8ta1
not modelled
8.0
31
Fold: Nucleotide-binding domainSuperfamily: Nucleotide-binding domainFamily: UDP-galactopyranose mutase, N-terminal domain78 d2bv3a5
not modelled
7.9
17
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: EF-G/eEF-2 domains III and V79 c2wghA_
not modelled
7.9
24
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase largePDBTitle: human ribonucleotide reductase r1 subunit (rrm1) in complex2 with datp and mg.
80 d2ezla_
not modelled
7.8
36
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain81 c4a1cV_
not modelled
7.6
16
PDB header: ribosomeChain: V: PDB Molecule: 60s ribosomal protein l7;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
82 c2zkr2_
not modelled
7.6
60
PDB header: ribosomal protein/rnaChain: 2: PDB Molecule: 60s ribosomal protein l37e;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
83 d2fyuk1
not modelled
7.6
39
Fold: Single transmembrane helixSuperfamily: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: Subunit XI (6.4 kDa protein) of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)84 c2cvuA_
not modelled
7.6
32
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase large chainPDBTitle: structures of yeast ribonucleotide reductase i
85 c3p5rB_
not modelled
7.4
23
PDB header: lyaseChain: B: PDB Molecule: taxadiene synthase;PDBTitle: crystal structure of taxadiene synthase from pacific yew (taxus2 brevifolia) in complex with mg2+ and 2-fluorogeranylgeranyl3 diphosphate
86 c3r1rB_
not modelled
7.4
28
PDB header: complex (oxidoreductase/peptide)Chain: B: PDB Molecule: ribonucleotide reductase r1 protein;PDBTitle: ribonucleotide reductase r1 protein with amppnp occupying2 the activity site from escherichia coli
87 c3jueA_
not modelled
7.4
32
PDB header: protein transport/endocytosisChain: A: PDB Molecule: arfgap with coiled-coil, ank repeat and ph domain-PDBTitle: crystal structure of arfgap and ank repeat domain of acap1
88 d2yt9a1
not modelled
7.3
63
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H289 c1s1iY_
not modelled
7.2
80
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l37-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
90 c3jywY_
not modelled
7.2
80
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l37(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
91 c2olvA_
not modelled
7.1
9
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding protein 2;PDBTitle: structural insight into the transglycosylation step of bacterial cell2 wall biosynthesis : donor ligand complex
92 d1b8da_
not modelled
7.1
28
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins93 c3fehA_
not modelled
7.1
38
PDB header: hydrolase activatorChain: A: PDB Molecule: centaurin-alpha-1;PDBTitle: crystal structure of full length centaurin alpha-1
94 c3k7bA_
not modelled
7.0
36
PDB header: viral proteinChain: A: PDB Molecule: protein a33;PDBTitle: the structure of the poxvirus a33 protein reveals a dimer of unique c-2 type lectin-like domains.
95 d1xp3a1
not modelled
6.9
22
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Endonuclease IV96 d1mm2a_
not modelled
6.9
31
Fold: FYVE/PHD zinc fingerSuperfamily: FYVE/PHD zinc fingerFamily: PHD domain97 d1bb8a_
not modelled
6.8
33
Fold: DNA-binding domainSuperfamily: DNA-binding domainFamily: DNA-binding domain from tn916 integrase98 d1zina2
not modelled
6.8
71
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain99 c1ybeA_
not modelled
6.7
23
PDB header: transferaseChain: A: PDB Molecule: nicotinate phosphoribosyltransferase;PDBTitle: crystal structure of a nicotinate phosphoribosyltransferase