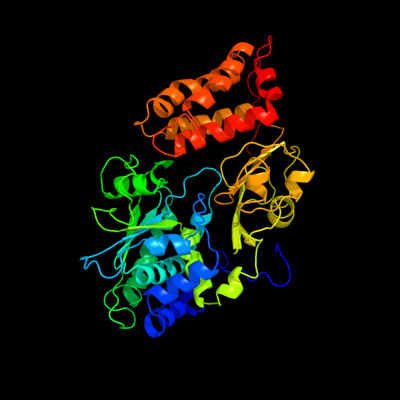
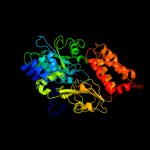
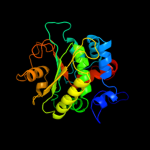
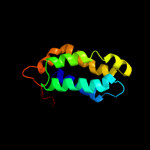
1 c2fugA_
100.0
46
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-quinone oxidoreductase chain 1;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
2 d2fug12
100.0
54
Fold: Nqo1 FMN-binding domain-likeSuperfamily: Nqo1 FMN-binding domain-likeFamily: Nqo1 FMN-binding domain-like3 d2fug11
100.0
32
Fold: Bromodomain-likeSuperfamily: Nqo1C-terminal domain-likeFamily: Nqo1C-terminal domain-like4 d2fug13
99.9
43
Fold: beta-Grasp (ubiquitin-like)Superfamily: Nqo1 middle domain-likeFamily: Nqo1 middle domain-like5 c2w8iG_
93.2
18
PDB header: membrane proteinChain: G: PDB Molecule: putative outer membrane lipoprotein wza;PDBTitle: crystal structure of wza24-345.
6 c2j58G_
91.9
18
PDB header: membrane proteinChain: G: PDB Molecule: outer membrane lipoprotein wza;PDBTitle: the structure of wza
7 c1c4cA_
78.1
11
PDB header: oxidoreductaseChain: A: PDB Molecule: protein (fe-only hydrogenase);PDBTitle: binding of exogenously added carbon monoxide at the active2 site of the fe-only hydrogenase (cpi) from clostridium3 pasteurianum
8 c3p42D_
65.8
13
PDB header: unknown functionChain: D: PDB Molecule: predicted protein;PDBTitle: structure of gfcc (ymcb), protein encoded by the e. coli group 42 capsule operon
9 d2bs2b1
46.1
20
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain10 c3swrA_
44.3
36
PDB header: transferaseChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 1;PDBTitle: structure of human dnmt1 (601-1600) in complex with sinefungin
11 c2b76N_
43.9
16
PDB header: oxidoreductaseChain: N: PDB Molecule: fumarate reductase iron-sulfur protein;PDBTitle: e. coli quinol fumarate reductase frda e49q mutation
12 c2bs2E_
42.1
20
PDB header: oxidoreductaseChain: E: PDB Molecule: quinol-fumarate reductase iron-sulfur subunit b;PDBTitle: quinol:fumarate reductase from wolinella succinogenes
13 d1kf6b1
41.3
17
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain14 c3bqqB_
37.7
7
PDB header: lipid binding proteinChain: B: PDB Molecule: proactivator polypeptide;PDBTitle: crystal structure of human saposin d (triclinic)
15 c1ffuA_
36.7
17
PDB header: hydrolaseChain: A: PDB Molecule: cuts, iron-sulfur protein of carbon monoxidePDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava which lacks the mo-pyranopterin moiety of the3 molybdenum cofactor
16 c2h89B_
36.0
18
PDB header: oxidoreductaseChain: B: PDB Molecule: succinate dehydrogenase ip subunit;PDBTitle: avian respiratory complex ii with malonate bound
17 c3bl5E_
35.9
19
PDB header: hydrolaseChain: E: PDB Molecule: queuosine biosynthesis protein quec;PDBTitle: crystal structure of quec from bacillus subtilis: an enzyme2 involved in preq1 biosynthesis
18 d1nkla_
35.3
12
Fold: Saposin-likeSuperfamily: SaposinFamily: NKL-like19 c3pt6B_
32.4
36
PDB header: transferase/dnaChain: B: PDB Molecule: dna (cytosine-5)-methyltransferase 1;PDBTitle: crystal structure of mouse dnmt1(650-1602) in complex with dna
20 d1zxia1
28.2
21
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like21 d1of9a_
not modelled
28.2
13
Fold: Saposin-likeSuperfamily: SaposinFamily: Ameobapore A22 c3thdD_
not modelled
25.2
26
PDB header: hydrolaseChain: D: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of human beta-galactosidase in complex with 1-2 deoxygalactonojirimycin
23 d1n62a1
not modelled
24.3
20
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like24 c3cf4A_
not modelled
23.2
23
PDB header: oxidoreductaseChain: A: PDB Molecule: acetyl-coa decarboxylase/synthase alpha subunit;PDBTitle: structure of the codh component of the m. barkeri acds complex
25 c1dgjA_
not modelled
22.6
14
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde oxidoreductase;PDBTitle: crystal structure of the aldehyde oxidoreductase from2 desulfovibrio desulfuricans atcc 27774
26 c3d3aA_
not modelled
22.3
26
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of a beta-galactosidase from bacteroides2 thetaiotaomicron
27 c1w59B_
not modelled
21.7
27
PDB header: cell divisionChain: B: PDB Molecule: cell division protein ftsz homolog 1;PDBTitle: ftsz dimer, empty (m. jannaschii)
28 c3b9jI_
not modelled
21.6
22
PDB header: oxidoreductaseChain: I: PDB Molecule: xanthine oxidase;PDBTitle: structure of xanthine oxidase with 2-hydroxy-6-methylpurine
29 d2gtga1
not modelled
20.0
12
Fold: Saposin-likeSuperfamily: SaposinFamily: NKL-like30 d1nekb1
not modelled
19.9
14
Fold: Globin-likeSuperfamily: alpha-helical ferredoxinFamily: Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain31 d1t3qa1
not modelled
18.7
21
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like32 d1v97a1
not modelled
18.4
22
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like33 c2he3A_
not modelled
18.0
9
PDB header: oxidoreductaseChain: A: PDB Molecule: glutathione peroxidase 2;PDBTitle: crystal structure of the selenocysteine to cysteine mutant of human2 glutathionine peroxidase 2 (gpx2)
34 d1o8xa_
not modelled
18.0
22
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like35 d1z6na1
not modelled
17.9
28
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Thioltransferase36 c1n60D_
not modelled
17.7
20
PDB header: oxidoreductaseChain: D: PDB Molecule: carbon monoxide dehydrogenase small chain;PDBTitle: crystal structure of the cu,mo-co dehydrogenase (codh); cyanide-2 inactivated form
37 c3eubJ_
not modelled
16.8
22
PDB header: oxidoreductaseChain: J: PDB Molecule: xanthine dehydrogenase/oxidase;PDBTitle: crystal structure of desulfo-xanthine oxidase with xanthine
38 d1ujsa_
not modelled
16.1
26
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain39 d2pg3a1
not modelled
16.1
44
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: N-type ATP pyrophosphatases40 d1deca_
not modelled
16.1
36
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Leech antihemostatic proteinsFamily: Hirudin-like41 c1nekB_
not modelled
15.9
18
PDB header: oxidoreductase/electron transportChain: B: PDB Molecule: succinate dehydrogenase iron-sulfur protein;PDBTitle: complex ii (succinate dehydrogenase) from e. coli with2 ubiquinone bound
42 d1l9la_
not modelled
15.6
6
Fold: Saposin-likeSuperfamily: SaposinFamily: NKL-like43 c1rm6F_
not modelled
15.5
20
PDB header: oxidoreductaseChain: F: PDB Molecule: 4-hydroxybenzoyl-coa reductase gamma subunit;PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
44 d1uufa1
not modelled
15.1
55
Fold: GroES-likeSuperfamily: GroES-likeFamily: Alcohol dehydrogenase-like, N-terminal domain45 d1i5ga_
not modelled
15.0
25
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like46 d1jqoa_
not modelled
14.8
35
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate carboxylase47 c1jqoA_
not modelled
14.8
35
PDB header: lyaseChain: A: PDB Molecule: phosphoenolpyruvate carboxylase;PDBTitle: crystal structure of c4-form phosphoenolpyruvate carboxylase from2 maize
48 c3nsjA_
not modelled
14.6
24
PDB header: immune systemChain: A: PDB Molecule: perforin-1;PDBTitle: the x-ray crystal structure of lymphocyte perforin
49 d1rm6c1
not modelled
14.2
18
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like50 c3o2qB_
not modelled
13.6
20
PDB header: hydrolaseChain: B: PDB Molecule: rna polymerase ii subunit a c-terminal domain phosphatasePDBTitle: crystal structure of the human symplekin-ssu72-ctd phosphopeptide2 complex
51 c3l23A_
not modelled
13.6
10
PDB header: isomeraseChain: A: PDB Molecule: sugar phosphate isomerase/epimerase;PDBTitle: crystal structure of sugar phosphate isomerase/epimerase2 (yp_001303399.1) from parabacteroides distasonis atcc 8503 at 1.70 a3 resolution
52 c2w3rG_
not modelled
13.4
15
PDB header: oxidoreductaseChain: G: PDB Molecule: xanthine dehydrogenase;PDBTitle: crystal structure of xanthine dehydrogenase (desulfo form)2 from rhodobacter capsulatus in complex with hypoxanthine
53 d1ejba_
not modelled
13.0
24
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase54 c2ds8A_
not modelled
12.9
35
PDB header: metal binding protein, protein bindingChain: A: PDB Molecule: atp-dependent clp protease atp-binding subunitPDBTitle: structure of the zbd-xb complex
55 c3mk3L_
not modelled
12.9
21
PDB header: transferaseChain: L: PDB Molecule: 6,7-dimethyl-8-ribityllumazine synthase;PDBTitle: crystal structure of lumazine synthase from salmonella typhimurium lt2
56 c3qmhA_
not modelled
12.8
64
PDB header: dna binding protein/dnaChain: A: PDB Molecule: cpg-binding protein;PDBTitle: structural basis of selective binding of non-methylated cpg islands2 (dna-tcga) by the cxxc domain of cfp1
57 d1c2ya_
not modelled
12.7
11
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase58 c2jyiA_
not modelled
12.7
64
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein hrx;PDBTitle: solution structure of mll cxxc domain
59 c3fdfA_
not modelled
12.5
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: fr253;PDBTitle: crystal structure of the serine phosphatase of rna2 polymerase ii ctd (ssu72 superfamily) from drosophila3 melanogaster. orthorhombic crystal form. northeast4 structural genomics consortium target fr253.
60 c3ogrA_
not modelled
12.1
27
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: complex structure of beta-galactosidase from trichoderma reesei with2 galactose
61 d1ffva1
not modelled
11.8
17
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like62 d1cdoa1
not modelled
11.8
36
Fold: GroES-likeSuperfamily: GroES-likeFamily: Alcohol dehydrogenase-like, N-terminal domain63 c1t3qD_
not modelled
11.7
21
PDB header: oxidoreductaseChain: D: PDB Molecule: quinoline 2-oxidoreductase small subunit;PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
64 c2l3xA_
not modelled
11.7
21
PDB header: protein bindingChain: A: PDB Molecule: ablim2 protein;PDBTitle: villin head piece domain of human ablim2
65 d1rvv1_
not modelled
11.7
17
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase66 c2bm0A_
not modelled
11.1
15
PDB header: elongation factorChain: A: PDB Molecule: elongation factor g;PDBTitle: ribosomal elongation factor g (ef-g) fusidic acid resistant2 mutant t84a
67 c2kvgA_
not modelled
11.0
26
PDB header: transcriptionChain: A: PDB Molecule: zinc finger and btb domain-containing protein 32;PDBTitle: structure of the three-cys2his2 domain of mouse testis zinc2 finger protein
68 d2fa8a1
not modelled
11.0
20
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Selenoprotein W-related69 c3hrdH_
not modelled
10.7
22
PDB header: oxidoreductaseChain: H: PDB Molecule: nicotinate dehydrogenase small fes subunit;PDBTitle: crystal structure of nicotinate dehydrogenase
70 c3p04B_
not modelled
10.6
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized bcr;PDBTitle: crystal structure of the bcr protein from corynebacterium glutamicum.2 northeast structural genomics consortium target cgr8
71 c2j2sA_
not modelled
10.6
64
PDB header: transcription regulationChain: A: PDB Molecule: zinc finger protein hrx;PDBTitle: solution structure of the nonmethyl-cpg-binding cxxc domain2 of the leukaemia-associated mll histone methyltransferase
72 d1yu5x1
not modelled
10.3
13
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain73 c2ojlB_
not modelled
10.2
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of q7waf1_borpa from bordetella parapertussis.2 northeast structural genomics target bpr68.
74 c1ovxB_
not modelled
10.2
35
PDB header: metal binding proteinChain: B: PDB Molecule: atp-dependent clp protease atp-binding subunit clpx;PDBTitle: nmr structure of the e. coli clpx chaperone zinc binding domain dimer
75 d2ds5a1
not modelled
10.2
35
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: ClpX chaperone zinc binding domain76 d1nqua_
not modelled
10.2
15
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase77 d2vapa1
not modelled
10.1
26
Fold: Tubulin nucleotide-binding domain-likeSuperfamily: Tubulin nucleotide-binding domain-likeFamily: Tubulin, GTPase domain78 c3o2sB_
not modelled
9.9
20
PDB header: hydrolaseChain: B: PDB Molecule: rna polymerase ii subunit a c-terminal domain phosphatasePDBTitle: crystal structure of the human symplekin-ssu72 complex
79 c2p0gB_
not modelled
9.7
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: selenoprotein w-related protein;PDBTitle: crystal structure of selenoprotein w-related protein from2 vibrio cholerae. northeast structural genomics target vcr75
80 c1xc6A_
not modelled
9.6
27
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: native structure of beta-galactosidase from penicillium sp. in complex2 with galactose
81 d1mmca_
not modelled
9.6
0
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Plant lectins/antimicrobial peptidesFamily: Antimicrobial peptide 2, AC-AMP282 c1ogpD_
not modelled
9.3
20
PDB header: oxidoreductaseChain: D: PDB Molecule: sulfite oxidase;PDBTitle: the crystal structure of plant sulfite oxidase provides2 insight into sulfite oxidation in plants and animals
83 d1lbua2
not modelled
8.9
12
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: Muramoyl-pentapeptide carboxypeptidase84 d1c41a_
not modelled
8.8
17
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase85 d1v9wa_
not modelled
8.8
28
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Txnl5-like86 d1o73a_
not modelled
8.7
27
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like87 c2obkE_
not modelled
8.5
20
PDB header: structural genomics, unknown functionChain: E: PDB Molecule: selt/selw/selh selenoprotein domain;PDBTitle: x-ray structure of the putative se binding protein from pseudomonas2 fluorescens. northeast structural genomics consortium target plr6.
88 c3p04A_
not modelled
8.5
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized bcr;PDBTitle: crystal structure of the bcr protein from corynebacterium glutamicum.2 northeast structural genomics consortium target cgr8
89 d1qzpa_
not modelled
8.5
18
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain90 c2h9gS_
not modelled
8.4
25
PDB header: immune system/apoptosisChain: S: PDB Molecule: tumor necrosis factor receptor superfamily member 10bPDBTitle: crystal structure of phage derived fab bdf1 with human death receptor2 5 (dr5)
91 c3fkfC_
not modelled
8.4
13
PDB header: oxidoreductaseChain: C: PDB Molecule: thiol-disulfide oxidoreductase;PDBTitle: thiol-disulfide oxidoreductase from bacteroides fragilis nctc 9343
92 c1zr6A_
not modelled
8.3
12
PDB header: oxidoreductaseChain: A: PDB Molecule: glucooligosaccharide oxidase;PDBTitle: the crystal structure of an acremonium strictum glucooligosaccharide2 oxidase reveals a novel flavinylation
93 c2obxH_
not modelled
8.1
15
PDB header: transferaseChain: H: PDB Molecule: 6,7-dimethyl-8-ribityllumazine synthase 1;PDBTitle: lumazine synthase ribh2 from mesorhizobium loti (gene mll7281, swiss-2 prot entry q986n2) complexed with inhibitor 5-nitro-6-(d-3 ribitylamino)-2,4(1h,3h) pyrimidinedione
94 c3dexA_
not modelled
8.1
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sav_2001;PDBTitle: crystal structure of sav_2001 protein from streptomyces2 avermitilis, northeast structural genomics consortium3 target svr107.
95 c1zlgA_
not modelled
8.0
31
PDB header: hormone/growth factorChain: A: PDB Molecule: anosmin 1;PDBTitle: solution structure of the extracellular matrix protein2 anosmin-1
96 d2fzwa1
not modelled
7.9
36
Fold: GroES-likeSuperfamily: GroES-likeFamily: Alcohol dehydrogenase-like, N-terminal domain97 d1di0a_
not modelled
7.8
17
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase98 c3l0aA_
not modelled
7.8
50
PDB header: hydrolaseChain: A: PDB Molecule: putative exonuclease;PDBTitle: crystal structure of putative exonuclease (rer070207002219) from2 eubacterium rectale at 2.19 a resolution
99 d2f9wa1
not modelled
7.7
23
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: CoaX-like