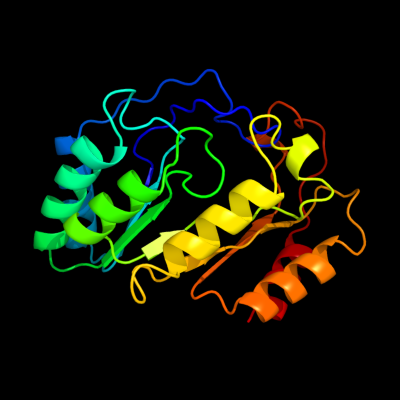
1 c3cixA_
99.9
20
PDB header: adomet binding proteinChain: A: PDB Molecule: fefe-hydrogenase maturase;PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
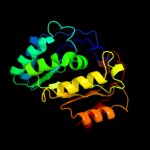
2 c1r30A_
99.9
17
PDB header: transferaseChain: A: PDB Molecule: biotin synthase;PDBTitle: the crystal structure of biotin synthase, an s-2 adenosylmethionine-dependent radical enzyme
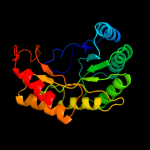
3 d1r30a_
99.9
17
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Biotin synthase4 c2qgqF_
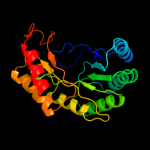
99.9
12
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: protein tm_1862;PDBTitle: crystal structure of tm_1862 from thermotoga maritima.2 northeast structural genomics consortium target vr77
5 c3t7vA_
99.9
15
PDB header: transferaseChain: A: PDB Molecule: methylornithine synthase pylb;PDBTitle: crystal structure of methylornithine synthase (pylb)
6 d1olta_
99.8
19
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: Oxygen-independent coproporphyrinogen III oxidase HemN7 d1tv8a_
99.8
13
Fold: TIM beta/alpha-barrelSuperfamily: Radical SAM enzymesFamily: MoCo biosynthesis proteins8 c3c8fA_
99.7
13
PDB header: oxidoreductaseChain: A: PDB Molecule: pyruvate formate-lyase 1-activating enzyme;PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with2 partially disordered adomet
9 c3rfaA_
99.6
13
PDB header: oxidoreductaseChain: A: PDB Molecule: ribosomal rna large subunit methyltransferase n;PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
10 c2yx0A_
99.5
16
PDB header: metal binding proteinChain: A: PDB Molecule: radical sam enzyme;PDBTitle: crystal structure of p. horikoshii tyw1
11 c2a5hC_
99.4
15
PDB header: isomeraseChain: C: PDB Molecule: l-lysine 2,3-aminomutase;PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
12 c3canA_
98.8
12
PDB header: lyase activatorChain: A: PDB Molecule: pyruvate-formate lyase-activating enzyme;PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
13 c2z2uA_
98.5
14
PDB header: metal binding proteinChain: A: PDB Molecule: upf0026 protein mj0257;PDBTitle: crystal structure of archaeal tyw1
14 c3ewbX_
97.8
13
PDB header: transferaseChain: X: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of n-terminal domain of putative 2-2 isopropylmalate synthase from listeria monocytogenes
15 c3ivuB_
97.7
12
PDB header: transferaseChain: B: PDB Molecule: homocitrate synthase, mitochondrial;PDBTitle: homocitrate synthase lys4 bound to 2-og
16 c3eegB_
97.6
7
PDB header: transferaseChain: B: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
17 c3bleA_
97.4
13
PDB header: transferaseChain: A: PDB Molecule: citramalate synthase from leptospira interrogans;PDBTitle: crystal structure of the catalytic domain of licms in2 complexed with malonate
18 c2ftpA_
97.4
13
PDB header: lyaseChain: A: PDB Molecule: hydroxymethylglutaryl-coa lyase;PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
19 d1nvma2
97.4
14
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: HMGL-like20 c2cw6B_
97.2
10
PDB header: lyaseChain: B: PDB Molecule: hydroxymethylglutaryl-coa lyase, mitochondrial;PDBTitle: crystal structure of human hmg-coa lyase: insights into2 catalysis and the molecular basis for3 hydroxymethylglutaric aciduria
21 c1rr2A_
not modelled
97.0
15
PDB header: transferaseChain: A: PDB Molecule: transcarboxylase 5s subunit;PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
22 c2nx9B_
not modelled
96.9
12
PDB header: lyaseChain: B: PDB Molecule: oxaloacetate decarboxylase 2, subunit alpha;PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
23 c1sr9A_
not modelled
96.9
12
PDB header: transferaseChain: A: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of leua from mycobacterium tuberculosis
24 c1nvmG_
not modelled
96.7
13
PDB header: lyase/oxidoreductaseChain: G: PDB Molecule: 4-hydroxy-2-oxovalerate aldolase;PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
25 c3navB_
not modelled
96.5
12
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
26 c2y7eA_
not modelled
96.2
19
PDB header: lyaseChain: A: PDB Molecule: 3-keto-5-aminohexanoate cleavage enzyme;PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
27 d1qopa_
not modelled
96.2
17
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes28 c1ydnA_
not modelled
96.0
9
PDB header: lyaseChain: A: PDB Molecule: hydroxymethylglutaryl-coa lyase;PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
29 c2ekcA_
not modelled
95.9
19
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
30 c3hpxB_
not modelled
95.9
14
PDB header: transferaseChain: B: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of mycobacterium tuberculosis leua active site2 domain 1-425 (truncation mutant delta:426-644)
31 c3no5C_
not modelled
95.8
16
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
32 c3c6cA_
not modelled
95.7
14
PDB header: hydrolaseChain: A: PDB Molecule: 3-keto-5-aminohexanoate cleavage enzyme;PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
33 c1ydoC_
not modelled
95.6
16
PDB header: lyaseChain: C: PDB Molecule: hmg-coa lyase;PDBTitle: crystal structure of the bacillis subtilis hmg-coa lyase, northeast2 structural genomics target sr181.
34 c3chvA_
not modelled
95.5
14
PDB header: metal binding proteinChain: A: PDB Molecule: prokaryotic domain of unknown function (duf849) with a timPDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
35 c2zyfA_
not modelled
95.3
10
PDB header: transferaseChain: A: PDB Molecule: homocitrate synthase;PDBTitle: crystal structure of homocitrate synthase from thermus thermophilus2 complexed with magnesuim ion and alpha-ketoglutarate
36 c3e02A_
not modelled
94.7
14
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein duf849;PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
37 d1rqba2
not modelled
94.4
15
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: HMGL-like38 c3b0vD_
not modelled
94.3
17
PDB header: oxidoreductase/rnaChain: D: PDB Molecule: trna-dihydrouridine synthase;PDBTitle: trna-dihydrouridine synthase from thermus thermophilus in complex with2 trna
39 d1b5ta_
not modelled
93.8
13
Fold: TIM beta/alpha-barrelSuperfamily: FAD-linked oxidoreductaseFamily: Methylenetetrahydrofolate reductase40 c3na8A_
not modelled
93.2
10
PDB header: lyaseChain: A: PDB Molecule: putative dihydrodipicolinate synthetase;PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
41 c3dxiB_
not modelled
93.1
7
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative aldolase;PDBTitle: crystal structure of the n-terminal domain of a putative2 aldolase (bvu_2661) from bacteroides vulgatus
42 c3khdC_
not modelled
92.9
10
PDB header: transferaseChain: C: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pff1300w.
43 c3ktcB_
not modelled
92.8
13
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
44 c3e49A_
not modelled
92.6
13
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein duf849 with a tim barrel fold;PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 with a tim barrel fold (bxe_c0966) from burkholderia xenovorans lb4003 at 1.75 a resolution
45 c2vc6A_
not modelled
92.5
12
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: structure of mosa from s. meliloti with pyruvate bound
46 c2ehhE_
not modelled
91.9
12
PDB header: lyaseChain: E: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
47 d1o5ka_
not modelled
91.9
14
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase48 d1yx1a1
91.7
14
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: KguE-like49 c3noeA_
not modelled
91.7
11
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
50 c3ggsA_
not modelled
91.6
15
PDB header: transferaseChain: A: PDB Molecule: purine nucleoside phosphorylase;PDBTitle: human purine nucleoside phosphorylase double mutant e201q,n243d2 complexed with 2-fluoro-2'-deoxyadenosine
51 c3lotC_
not modelled
91.5
19
PDB header: structure genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein of unknown function (np_070038.1) from2 archaeoglobus fulgidus at 1.89 a resolution
52 c3lciA_
not modelled
91.4
14
PDB header: lyaseChain: A: PDB Molecule: n-acetylneuraminate lyase;PDBTitle: the d-sialic acid aldolase mutant v251w
53 c3g0sA_
not modelled
91.0
13
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
54 c3n2xB_
not modelled
90.9
12
PDB header: lyaseChain: B: PDB Molecule: uncharacterized protein yage;PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
55 c3daqB_
not modelled
90.8
11
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from methicillin-2 resistant staphylococcus aureus
56 c2r8wB_
not modelled
90.8
15
PDB header: lyaseChain: B: PDB Molecule: agr_c_1641p;PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
57 c3labA_
not modelled
90.7
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative kdpg (2-keto-3-deoxy-6-phosphogluconate)PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
58 c2yxgD_
not modelled
90.7
13
PDB header: lyaseChain: D: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
59 c2pa6A_
not modelled
90.7
14
PDB header: lyaseChain: A: PDB Molecule: enolase;PDBTitle: crystal structure of mj0232 from methanococcus jannaschii
60 c3eb2A_
not modelled
90.6
12
PDB header: lyaseChain: A: PDB Molecule: putative dihydrodipicolinate synthetase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
61 c3lerA_
not modelled
90.5
14
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
62 c3bi8A_
not modelled
90.3
10
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
63 c3c52B_
not modelled
90.2
9
PDB header: lyaseChain: B: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: class ii fructose-1,6-bisphosphate aldolase from2 helicobacter pylori in complex with3 phosphoglycolohydroxamic acid, a competitive inhibitor
64 c2rfgB_
not modelled
90.2
11
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
65 c2v9dB_
not modelled
90.1
12
PDB header: lyaseChain: B: PDB Molecule: yage;PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
66 d1f74a_
not modelled
90.1
11
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase67 d3pnpa_
not modelled
89.9
18
Fold: Phosphorylase/hydrolase-likeSuperfamily: Purine and uridine phosphorylasesFamily: Purine and uridine phosphorylases68 c2f06B_
not modelled
89.8
10
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
69 d1xkya1
not modelled
89.7
14
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase70 d1xxxa1
not modelled
89.7
16
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase71 d2a6na1
not modelled
89.5
12
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase72 c3fluD_
not modelled
89.4
13
PDB header: lyaseChain: D: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
73 d3bgsa1
not modelled
89.4
16
Fold: Phosphorylase/hydrolase-likeSuperfamily: Purine and uridine phosphorylasesFamily: Purine and uridine phosphorylases74 d1hl2a_
not modelled
89.3
14
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase75 c2fmoA_
not modelled
89.3
14
PDB header: oxidoreductaseChain: A: PDB Molecule: 5,10-methylenetetrahydrofolate reductase;PDBTitle: ala177val mutant of e. coli methylenetetrahydrofolate2 reductase
76 c2pjuD_
not modelled
88.7
12
PDB header: transcriptionChain: D: PDB Molecule: propionate catabolism operon regulatory protein;PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
77 c3q94B_
not modelled
88.4
10
PDB header: lyaseChain: B: PDB Molecule: fructose-bisphosphate aldolase, class ii;PDBTitle: the crystal structure of fructose 1,6-bisphosphate aldolase from2 bacillus anthracis str. 'ames ancestor'
78 c2qv5A_
not modelled
88.4
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu2773;PDBTitle: crystal structure of uncharacterized protein atu2773 from2 agrobacterium tumefaciens c58
79 c3pueA_
not modelled
88.4
11
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
80 c3si9B_
not modelled
88.3
11
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from bartonella2 henselae
81 c3kwsB_
not modelled
88.1
9
PDB header: isomeraseChain: B: PDB Molecule: putative sugar isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
82 c3qfeB_
not modelled
87.6
14
PDB header: lyaseChain: B: PDB Molecule: putative dihydrodipicolinate synthase family protein;PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
83 c3h5dD_
not modelled
87.6
18
PDB header: lyaseChain: D: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: dihydrodipicolinate synthase from drug-resistant streptococcus2 pneumoniae
84 c3g8rA_
not modelled
87.5
12
PDB header: biosynthetic proteinChain: A: PDB Molecule: probable spore coat polysaccharide biosynthesis protein e;PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
85 d2pjua1
not modelled
87.1
12
Fold: Chelatase-likeSuperfamily: PrpR receptor domain-likeFamily: PrpR receptor domain-like86 d1mxsa_
not modelled
86.9
20
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase87 d1ps9a1
not modelled
86.6
15
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases88 d1pkla2
not modelled
86.6
14
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase89 c3e96B_
not modelled
86.5
13
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
90 d1liua2
not modelled
86.2
13
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase91 c3cprB_
not modelled
86.0
15
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthetase;PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
92 c3otrC_
not modelled
85.9
13
PDB header: lyaseChain: C: PDB Molecule: enolase;PDBTitle: 2.75 angstrom crystal structure of enolase 1 from toxoplasma gondii
93 d1xcfa_
not modelled
85.3
17
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes94 d1k77a_
not modelled
84.8
12
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Hypothetical protein YgbM (EC1530)95 d1muma_
not modelled
84.8
17
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Phosphoenolpyruvate mutase/Isocitrate lyase-like96 c2r94B_
not modelled
84.2
13
PDB header: lyaseChain: B: PDB Molecule: 2-keto-3-deoxy-(6-phospho-)gluconate aldolase;PDBTitle: crystal structure of kd(p)ga from t.tenax
97 d1oy0a_
not modelled
83.8
16
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Ketopantoate hydroxymethyltransferase PanB98 c3d0cB_
not modelled
83.5
15
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 oceanobacillus iheyensis at 1.9 a resolution
99 d1wbha1
not modelled
83.4
19
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase100 c3s5oA_
not modelled
83.1
17
PDB header: lyaseChain: A: PDB Molecule: 4-hydroxy-2-oxoglutarate aldolase, mitochondrial;PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
101 d1vhca_
not modelled
82.6
20
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase102 d1vp8a_
not modelled
82.1
15
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: MTH1675-like103 d1thfd_
not modelled
81.9
14
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Histidine biosynthesis enzymes104 c3kw2A_
not modelled
81.5
10
PDB header: transferaseChain: A: PDB Molecule: probable r-rna methyltransferase;PDBTitle: crystal structure of probable rrna-methyltransferase from2 porphyromonas gingivalis
105 c3fkkA_
80.6
13
PDB header: lyaseChain: A: PDB Molecule: l-2-keto-3-deoxyarabonate dehydratase;PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
106 c3ct7E_
not modelled
80.6
10
PDB header: isomeraseChain: E: PDB Molecule: d-allulose-6-phosphate 3-epimerase;PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
107 d1i60a_
not modelled
80.3
11
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like108 c3bg3B_
not modelled
79.7
16
PDB header: ligaseChain: B: PDB Molecule: pyruvate carboxylase, mitochondrial;PDBTitle: crystal structure of human pyruvate carboxylase (missing2 the biotin carboxylase domain at the n-terminus)
109 d1gvfa_
not modelled
79.5
8
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class II FBP aldolase110 c3aamA_
not modelled
79.4
15
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease iv;PDBTitle: crystal structure of endonuclease iv from thermus thermophilus hb8
111 c3dz1A_
not modelled
78.6
13
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
112 c1zlpA_
not modelled
78.5
18
PDB header: lyaseChain: A: PDB Molecule: petal death protein;PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
113 c2z1kA_
not modelled
78.4
23
PDB header: hydrolaseChain: A: PDB Molecule: (neo)pullulanase;PDBTitle: crystal structure of ttha1563 from thermus thermophilus hb8
114 d2akza1
not modelled
77.9
15
Fold: TIM beta/alpha-barrelSuperfamily: Enolase C-terminal domain-likeFamily: Enolase115 c1jpkA_
not modelled
77.8
12
PDB header: lyaseChain: A: PDB Molecule: uroporphyrinogen decarboxylase;PDBTitle: gly156asp mutant of human urod, human uroporphyrinogen iii2 decarboxylase
116 c3dx5A_
not modelled
77.7
11
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein asbf;PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
117 c3b4uB_
not modelled
77.2
15
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
118 d1ktba2
not modelled
77.2
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain119 c1xuzA_
not modelled
76.4
20
PDB header: biosynthetic proteinChain: A: PDB Molecule: polysialic acid capsule biosynthesis protein siac;PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
120 c1uasA_
not modelled
76.2
10
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase;PDBTitle: crystal structure of rice alpha-galactosidase