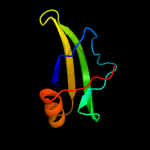
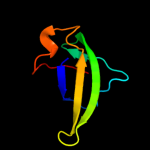
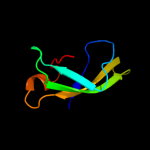
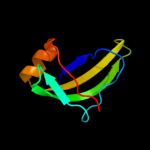
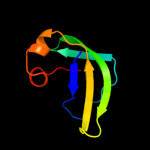
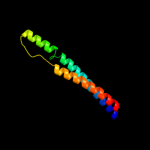
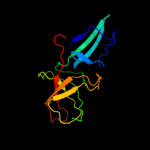
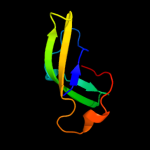
1 d1pj5a1
93.4
17
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain2 d1v5va1
90.3
17
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain3 d1wosa1
87.9
17
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain4 c1v5vA_
84.2
17
PDB header: transferaseChain: A: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of a component of glycine cleavage system: t-protein2 from pyrococcus horikoshii ot3 at 1.5 a resolution
5 c1qu7A_
78.6
9
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein i;PDBTitle: four helical-bundle structure of the cytoplasmic domain of a serine2 chemotaxis receptor
6 c3girA_
75.4
15
PDB header: transferaseChain: A: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of glycine cleavage system2 aminomethyltransferase t from bartonella henselae
7 c1worA_
75.3
18
PDB header: transferaseChain: A: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of t-protein of the glycine cleavage2 system
8 c2wpqA_
74.7
5
PDB header: membrane proteinChain: A: PDB Molecule: trimeric autotransporter adhesin fragment;PDBTitle: salmonella enterica sada 479-519 fused to gcn4 adaptors (2 sadak3, in-register fusion)
9 c2j5uB_
73.9
16
PDB header: cell shape regulationChain: B: PDB Molecule: mrec protein;PDBTitle: mrec lysteria monocytogenes
10 c1ei3C_
72.9
13
PDB header: PDB COMPND: 11 c1yx2B_
72.6
23
PDB header: transferaseChain: B: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of the probable aminomethyltransferase2 from bacillus subtilis
12 c1pj6A_
68.4
17
PDB header: oxidoreductaseChain: A: PDB Molecule: n,n-dimethylglycine oxidase;PDBTitle: crystal structure of dimethylglycine oxidase of arthrobacter2 globiformis in complex with folic acid
13 c2ieqC_
67.9
17
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: core structure of s2 from the human coronavirus nl63 spike2 glycoprotein
14 c2qf4A_
64.9
15
PDB header: structural proteinChain: A: PDB Molecule: cell shape determining protein mrec;PDBTitle: high resolution structure of the major periplasmic domain from the2 cell shape-determining filament mrec (orthorhombic form)
15 d1vloa1
63.6
18
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain16 c3tfhB_
58.3
14
PDB header: transferaseChain: B: PDB Molecule: gcvt-like aminomethyltransferase protein;PDBTitle: dmsp-dependent demethylase from p. ubique - apo
17 d1c99a_
57.5
28
Fold: Transmembrane helix hairpinSuperfamily: F1F0 ATP synthase subunit CFamily: F1F0 ATP synthase subunit C18 c2d4yA_
53.0
11
PDB header: structural proteinChain: A: PDB Molecule: flagellar hook-associated protein 1;PDBTitle: crystal structure of a 49k fragment of hap1 (flgk)
19 c3g67A_
51.8
5
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of a soluble chemoreceptor from thermotoga2 maritima
20 d1txka2
50.8
24
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: MdoG-like21 c1kmiZ_
not modelled
49.0
12
PDB header: signaling proteinChain: Z: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of an e.coli chemotaxis protein, chez
22 c1wsrA_
not modelled
48.0
15
PDB header: transferaseChain: A: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of human t-protein of glycine cleavage2 system
23 d1fftb2
not modelled
46.5
21
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region24 c1txkA_
not modelled
46.2
24
PDB header: biosynthetic proteinChain: A: PDB Molecule: glucans biosynthesis protein g;PDBTitle: crystal structure of escherichia coli opgg
25 c3lnrA_
not modelled
40.7
13
PDB header: signaling proteinChain: A: PDB Molecule: aerotaxis transducer aer2;PDBTitle: crystal structure of poly-hamp domains from the p. aeruginosa soluble2 receptor aer2
26 d1vp7b_
not modelled
36.6
17
Fold: Spectrin repeat-likeSuperfamily: XseB-likeFamily: XseB-like27 c2ch7A_
not modelled
35.6
7
PDB header: chemotaxisChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of the cytoplasmic domain of a bacterial2 chemoreceptor from thermotoga maritima
28 c1wyyB_
not modelled
35.1
15
PDB header: viral proteinChain: B: PDB Molecule: e2 glycoprotein;PDBTitle: post-fusion hairpin conformation of the sars coronavirus spike2 glycoprotein
29 c1ei3E_
not modelled
35.0
9
PDB header: PDB COMPND: 30 c1deqF_
not modelled
31.7
6
PDB header: PDB COMPND: 31 c2vs0B_
not modelled
31.2
9
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
32 d1jz8a3
not modelled
30.3
11
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain33 c3gvmA_
not modelled
30.1
8
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
34 d1eq1a_
not modelled
29.0
10
Fold: Apolipophorin-IIISuperfamily: Apolipophorin-IIIFamily: Apolipophorin-III35 d1yq2a3
not modelled
27.6
15
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain36 d1szia_
not modelled
27.3
11
Fold: Four-helical up-and-down bundleSuperfamily: Mannose-6-phosphate receptor binding protein 1 (Tip47), C-terminal domainFamily: Mannose-6-phosphate receptor binding protein 1 (Tip47), C-terminal domain37 c1kqsA_
not modelled
27.0
19
PDB header: ribosomeChain: A: PDB Molecule: ribosomal protein l2;PDBTitle: the haloarcula marismortui 50s complexed with a2 pretranslocational intermediate in protein synthesis
38 d1ucua_
not modelled
26.9
8
Fold: Phase 1 flagellinSuperfamily: Phase 1 flagellinFamily: Phase 1 flagellin39 d1g4us1
not modelled
25.6
19
Fold: Four-helical up-and-down bundleSuperfamily: Bacterial GAP domainFamily: Bacterial GAP domain40 d1v58a2
not modelled
25.5
17
Fold: Cystatin-likeSuperfamily: DsbC/DsbG N-terminal domain-likeFamily: DsbC/DsbG N-terminal domain-like41 c1deqO_
not modelled
24.7
14
PDB header: PDB COMPND: 42 c2kebA_
not modelled
24.6
21
PDB header: dna binding proteinChain: A: PDB Molecule: dna polymerase subunit alpha b;PDBTitle: nmr solution structure of the n-terminal domain of the dna polymerase2 alpha p68 subunit
43 c3k8wA_
not modelled
22.6
10
PDB header: structural proteinChain: A: PDB Molecule: flagellin homolog;PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c45
44 c3izcN_
not modelled
22.4
16
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein rpl14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
45 c2kbbA_
not modelled
21.9
11
PDB header: structural proteinChain: A: PDB Molecule: talin-1;PDBTitle: nmr structure of the talin rod domain, 1655-1822
46 d1im3d_
not modelled
21.5
22
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Cytomegalovirus protein US247 d2axte1
not modelled
21.3
15
Fold: Single transmembrane helixSuperfamily: Cytochrome b559 subunitsFamily: Cytochrome b559 subunits48 c2d4xA_
not modelled
21.3
6
PDB header: structural proteinChain: A: PDB Molecule: flagellar hook-associated protein 3;PDBTitle: crystal structure of a 26k fragment of hap3 (flgl)
49 d1gtra1
not modelled
21.0
16
Fold: Ribosomal protein L25-likeSuperfamily: Ribosomal protein L25-likeFamily: Gln-tRNA synthetase (GlnRS), C-terminal (anticodon-binding) domain50 c2nrjA_
not modelled
21.0
15
PDB header: toxinChain: A: PDB Molecule: hbl b protein;PDBTitle: crystal structure of hemolysin binding component from2 bacillus cereus
51 d1lvfa_
not modelled
20.4
11
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins52 c2gl2B_
not modelled
20.2
14
PDB header: cell adhesionChain: B: PDB Molecule: adhesion a;PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,2 y69g) of bacterial adhesin fada
53 c3ghgK_
not modelled
18.4
8
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
54 c1x31A_
not modelled
18.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: sarcosine oxidase alpha subunit;PDBTitle: crystal structure of heterotetrameric sarcosine oxidase from2 corynebacterium sp. u-96
55 c3o0lB_
not modelled
18.1
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a pfam duf1425 family member (shew_1734) from2 shewanella sp. pv-4 at 1.81 a resolution
56 d1dowa_
not modelled
17.9
11
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin57 c4a19F_
not modelled
17.4
19
PDB header: ribosomeChain: F: PDB Molecule: rpl14;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 2.
58 c3k8vB_
not modelled
17.0
10
PDB header: structural proteinChain: B: PDB Molecule: flagellin homolog;PDBTitle: crysatl structure of a bacterial cell-surface flagellin n20c20
59 d1wrda1
not modelled
16.9
18
Fold: Spectrin repeat-likeSuperfamily: GAT-like domainFamily: GAT domain60 d2zjrs1
not modelled
16.3
20
Fold: Ribosomal protein L25-likeSuperfamily: Ribosomal protein L25-likeFamily: Ribosomal protein L25-like61 d1bhua_
not modelled
16.1
23
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Streptomyces metalloproteinase inhibitor, SMPI62 d1dova_
not modelled
15.5
11
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin63 c1vloA_
not modelled
15.4
18
PDB header: transferaseChain: A: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of aminomethyltransferase (t protein;2 tetrahydrofolate-dependent) of glycine cleavage system (np417381)3 from escherichia coli k12 at 1.70 a resolution
64 c3o5aB_
not modelled
15.4
18
PDB header: oxidoreductaseChain: B: PDB Molecule: diheme cytochrome c napb;PDBTitle: crystal structure of partially reduced periplasmic nitrate reductase2 from cupriavidus necator using ionic liquids
65 c1rkeA_
not modelled
14.9
10
PDB header: cell adhesion, structural proteinChain: A: PDB Molecule: vinculin;PDBTitle: human vinculin head (1-258) in complex with human vinculin2 tail (879-1066)
66 c3ojaB_
not modelled
14.8
10
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
67 d2cp5a1
not modelled
14.7
11
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain68 d1bhga2
not modelled
14.6
16
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain69 c3ls1A_
not modelled
13.4
18
PDB header: photosynthesisChain: A: PDB Molecule: sll1638 protein;PDBTitle: crystal structure of cyanobacterial psbq from synechocystis2 sp. pcc 6803 complexed with zn2+
70 d1pa4a_
not modelled
13.0
44
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA71 d1feua_
not modelled
13.0
14
Fold: Ribosomal protein L25-likeSuperfamily: Ribosomal protein L25-likeFamily: Ribosomal protein L25-like72 c3iz5N_
not modelled
12.8
14
PDB header: ribosomeChain: N: PDB Molecule: 60s ribosomal protein l14 (l14e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
73 c2dq3A_
not modelled
12.8
11
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: crystal structure of aq_298
74 c2kncA_
not modelled
12.6
35
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
75 c1vlyA_
not modelled
12.6
10
PDB header: transferaseChain: A: PDB Molecule: unknown protein from 2d-page;PDBTitle: crystal structure of a putative aminomethyltransferase (ygfz) from2 escherichia coli at 1.30 a resolution
76 c1zvzA_
not modelled
12.6
9
PDB header: protein bindingChain: A: PDB Molecule: vinculin;PDBTitle: vinculin head (0-258) in complex with the talin rod residue2 820-844
77 d2cp2a1
not modelled
11.9
16
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain78 d1kkga_
not modelled
11.8
19
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA79 c3nmbA_
not modelled
11.0
64
PDB header: hydrolaseChain: A: PDB Molecule: putative sugar hydrolase;PDBTitle: crystal structure of a putative sugar hydrolase (bacova_03189) from2 bacteroides ovatus at 2.40 a resolution
80 d1wa8a1
not modelled
10.8
11
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like81 d1qoua_
not modelled
10.5
11
Fold: PEBP-likeSuperfamily: PEBP-likeFamily: Phosphatidylethanolamine binding protein82 c3hd7A_
not modelled
10.4
8
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
83 d2coza1
not modelled
10.4
14
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain84 d1sj8a2
not modelled
10.4
10
Fold: I/LWEQ domainSuperfamily: I/LWEQ domainFamily: I/LWEQ domain85 c1exdA_
not modelled
10.3
19
PDB header: ligase/rnaChain: A: PDB Molecule: glutaminyl-trna synthetase;PDBTitle: crystal structure of a tight-binding glutamine trna bound2 to glutamine aminoacyl trna synthetase
86 c1m57H_
not modelled
10.1
18
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c oxidase;PDBTitle: structure of cytochrome c oxidase from rhodobacter2 sphaeroides (eq(i-286) mutant))
87 c3dshA_
not modelled
10.0
17
PDB header: dna binding proteinChain: A: PDB Molecule: interferon regulatory factor 5;PDBTitle: crystal structure of dimeric interferon regulatory factor 5 (irf-5)2 transactivation domain
88 c1urqC_
not modelled
9.9
14
PDB header: transport proteinChain: C: PDB Molecule: synaptosomal-associated protein 25;PDBTitle: crystal structure of neuronal q-snares in complex with2 r-snare motif of tomosyn
89 d1t01a1
not modelled
9.9
15
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin90 c3lw9B_
not modelled
9.8
24
PDB header: protein transportChain: B: PDB Molecule: invasion protein inva;PDBTitle: structure of a cytoplasmic domain of salmonella inva
91 c1zoqA_
not modelled
9.8
25
PDB header: transcription/transferaseChain: A: PDB Molecule: interferon regulatory factor 3;PDBTitle: irf3-cbp complex
92 c2kzfA_
not modelled
9.7
13
PDB header: ribosomal proteinChain: A: PDB Molecule: ribosome-binding factor a;PDBTitle: solution nmr structure of the thermotoga maritima protein tm0855 a2 putative ribosome binding factor a
93 c3h3lB_
not modelled
9.7
38
PDB header: hydrolaseChain: B: PDB Molecule: putative sugar hydrolase;PDBTitle: crystal structure of putative sugar hydrolase (yp_001304206.1) from2 parabacteroides distasonis atcc 8503 at 1.59 a resolution
94 c2dnxA_
not modelled
9.6
16
PDB header: transport proteinChain: A: PDB Molecule: syntaxin-12;PDBTitle: solution structure of rsgi ruh-063, an n-terminal domain of2 syntaxin 12 from human cdna
95 d1wgba_
not modelled
9.5
13
Fold: Split barrel-likeSuperfamily: FMN-binding split barrelFamily: NADH:FMN oxidoreductase-like96 c3hbkA_
not modelled
9.4
55
PDB header: hydrolaseChain: A: PDB Molecule: putative glycosyl hydrolase;PDBTitle: crystal structure of putative glycosyl hydrolase, was domain of2 unknown function (duf1080) (yp_001302580.1) from parabacteroides3 distasonis atcc 8503 at 2.36 a resolution
97 c1cn3F_
not modelled
9.4
25
PDB header: viral proteinChain: F: PDB Molecule: fragment of coat protein vp2;PDBTitle: interaction of polyomavirus internal protein vp2 with major2 capsid protein vp1 and implications for participation of3 vp2 in viral entry
98 c1pkvB_
not modelled
9.4
13
PDB header: transferaseChain: B: PDB Molecule: riboflavin synthase alpha chain;PDBTitle: the n-terminal domain of riboflavin synthase in complex with2 riboflavin
99 c1pkvA_
not modelled
9.4
13
PDB header: transferaseChain: A: PDB Molecule: riboflavin synthase alpha chain;PDBTitle: the n-terminal domain of riboflavin synthase in complex with2 riboflavin