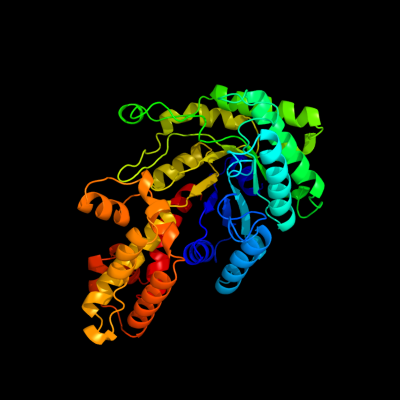
| 1 | d2fiqa1
|
|
 |
100.0 |
53 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:GatZ-like |
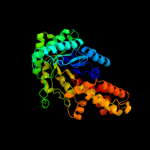
| 2 | c3q94B_
|
|
 |
99.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase, class ii;
PDBTitle: the crystal structure of fructose 1,6-bisphosphate aldolase from2 bacillus anthracis str. 'ames ancestor'
|
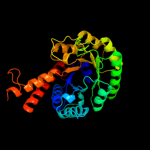
| 3 | d1rvga_
|
|
 |
99.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
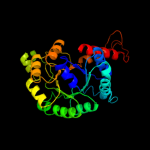
| 4 | d1dosa_
|
|
 |
99.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 5 | d1gvfa_
|
|
 |
99.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 6 | c3qm3C_
|
|
 |
99.8 |
18 |
PDB header:lyase
Chain: C: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: 1.85 angstrom resolution crystal structure of fructose-bisphosphate2 aldolase (fba) from campylobacter jejuni
|
| 7 | c3c52B_
|
|
 |
99.8 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: class ii fructose-1,6-bisphosphate aldolase from2 helicobacter pylori in complex with3 phosphoglycolohydroxamic acid, a competitive inhibitor
|
| 8 | c3pm6B_
|
|
 |
99.8 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-bisphosphate aldolase;
PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
|
| 9 | c3elfA_
|
|
 |
99.8 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: structural characterization of tetrameric mycobacterium tuberculosis2 fructose 1,6-bisphosphate aldolase - substrate binding and catalysis3 mechanism of a class iia bacterial aldolase
|
| 10 | c2iswB_
|
|
 |
99.8 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-1,6-bisphosphate aldolase;
PDBTitle: structure of giardia fructose-1,6-biphosphate aldolase in2 complex with phosphoglycolohydroxamate
|
| 11 | d1f6ya_
|
|
 |
95.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 12 | c3bolB_
|
|
 |
94.8 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
| 13 | d3bofa1
|
|
 |
93.6 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 14 | c1rr2A_
|
|
 |
93.2 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
| 15 | c3k13A_
|
|
 |
92.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
|
| 16 | c2yciX_
|
|
 |
91.6 |
14 |
PDB header:transferase
Chain: X: PDB Molecule:5-methyltetrahydrofolate corrinoid/iron sulfur protein
PDBTitle: methyltransferase native
|
| 17 | c2h9aA_
|
|
 |
91.6 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carbon monoxide dehydrogenase corrinoid/iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 18 | c2nx9B_
|
|
 |
91.3 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:oxaloacetate decarboxylase 2, subunit alpha;
PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
|
| 19 | c3cqkB_
|
|
 |
87.7 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
| 20 | c3k2gA_
|
|
 |
86.7 |
21 |
PDB header:resiniferatoxin binding protein
Chain: A: PDB Molecule:resiniferatoxin-binding, phosphotriesterase-
PDBTitle: crystal structure of a resiniferatoxin-binding protein from2 rhodobacter sphaeroides
|
| 21 | c2vp8A_ |
|
not modelled |
86.5 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase 2;
PDBTitle: structure of mycobacterium tuberculosis rv1207
|
| 22 | c2dzaA_ |
|
not modelled |
85.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
| 23 | c3lhlA_ |
|
not modelled |
85.1 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative agmatinase;
PDBTitle: crystal structure of a putative agmatinase from clostridium difficile
|
| 24 | c2ze3A_ |
|
not modelled |
83.6 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 25 | d1eyea_ |
|
not modelled |
82.4 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 26 | d1muma_ |
|
not modelled |
81.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 27 | d1ad1a_ |
|
not modelled |
81.3 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 28 | c2uvaI_ |
|
not modelled |
79.4 |
25 |
PDB header:transferase
Chain: I: PDB Molecule:fatty acid synthase beta subunits;
PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the beta subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
|
| 29 | d2a0ma1 |
|
not modelled |
79.0 |
20 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 30 | d1xfka_ |
|
not modelled |
77.8 |
20 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 31 | c3o1lB_ |
|
not modelled |
77.3 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
|
| 32 | d2ceva_ |
|
not modelled |
77.1 |
24 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 33 | c2vkzH_ |
|
not modelled |
76.9 |
20 |
PDB header:transferase
Chain: H: PDB Molecule:fatty acid synthase subunit beta;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid2 synthase type i multienzyme complex
|
| 34 | d1gq6a_ |
|
not modelled |
76.4 |
18 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 35 | c3nioF_ |
|
not modelled |
76.3 |
20 |
PDB header:hydrolase
Chain: F: PDB Molecule:guanidinobutyrase;
PDBTitle: crystal structure of pseudomonas aeruginosa guanidinobutyrase
|
| 36 | c3nrbD_ |
|
not modelled |
76.0 |
16 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
| 37 | d1vhna_ |
|
not modelled |
75.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 38 | d1t4ba1 |
|
not modelled |
74.2 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 39 | d1pq3a_ |
|
not modelled |
73.0 |
17 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 40 | c2y5sA_ |
|
not modelled |
70.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
| 41 | d1ajza_ |
|
not modelled |
69.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 42 | d1tx2a_ |
|
not modelled |
67.6 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 43 | c1tx2A_ |
|
not modelled |
67.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 44 | c2hjpA_ |
|
not modelled |
66.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
| 45 | c3obiC_ |
|
not modelled |
66.0 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
| 46 | c2qiwA_ |
|
not modelled |
66.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
| 47 | c3nipB_ |
|
not modelled |
65.5 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:3-guanidinopropionase;
PDBTitle: crystal structure of pseudomonas aeruginosa guanidinopropionase2 complexed with 1,6-diaminohexane
|
| 48 | d1woha_ |
|
not modelled |
64.6 |
20 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 49 | c3mmrA_ |
|
not modelled |
62.8 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:arginase;
PDBTitle: structure of plasmodium falciparum arginase in complex with abh
|
| 50 | c3louB_ |
|
not modelled |
62.4 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
| 51 | c3n0vD_ |
|
not modelled |
62.2 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 52 | d1aw1a_ |
|
not modelled |
61.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 53 | c1vraB_ |
|
not modelled |
58.4 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:arginine biosynthesis bifunctional protein argj;
PDBTitle: crystal structure of arginine biosynthesis bifunctional protein argj2 (10175521) from bacillus halodurans at 2.00 a resolution
|
| 54 | c1ydnA_ |
|
not modelled |
58.3 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
| 55 | c3it4B_ |
|
not modelled |
58.1 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:arginine biosynthesis bifunctional protein argj
PDBTitle: the crystal structure of ornithine acetyltransferase from2 mycobacterium tuberculosis (rv1653) at 1.7 a
|
| 56 | d2ihta3 |
|
not modelled |
56.6 |
31 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
| 57 | c3r4iB_ |
|
not modelled |
56.3 |
4 |
PDB header:lyase
Chain: B: PDB Molecule:citrate lyase;
PDBTitle: crystal structure of a citrate lyase (bxe_b2899) from burkholderia2 xenovorans lb400 at 2.24 a resolution
|
| 58 | c3lerA_ |
|
not modelled |
55.1 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
|
| 59 | c1cqxB_ |
|
not modelled |
55.0 |
19 |
PDB header:lipid binding protein
Chain: B: PDB Molecule:flavohemoprotein;
PDBTitle: crystal structure of the flavohemoglobin from alcaligenes eutrophus at2 1.75 a resolution
|
| 60 | d1pqua1 |
|
not modelled |
54.7 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 61 | d2aeba1 |
|
not modelled |
52.4 |
22 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 62 | c3pzlA_ |
|
not modelled |
51.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:agmatine ureohydrolase;
PDBTitle: the crystal structure of agmatine ureohydrolase of thermoplasma2 volcanium
|
| 63 | c3d0kA_ |
|
not modelled |
50.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative poly(3-hydroxybutyrate) depolymerase lpqc;
PDBTitle: crystal structure of the lpqc, poly(3-hydroxybutyrate) depolymerase2 from bordetella parapertussis
|
| 64 | d1mb4a1 |
|
not modelled |
49.7 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 65 | c3dxiB_ |
|
not modelled |
48.8 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative aldolase;
PDBTitle: crystal structure of the n-terminal domain of a putative2 aldolase (bvu_2661) from bacteroides vulgatus
|
| 66 | d1yl7a1 |
|
not modelled |
48.5 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 67 | c3m1rF_ |
|
not modelled |
48.3 |
23 |
PDB header:hydrolase
Chain: F: PDB Molecule:formimidoylglutamase;
PDBTitle: the crystal structure of formimidoylglutamase from bacillus2 subtilis subsp. subtilis str. 168
|
| 68 | d1ujqa_ |
|
not modelled |
48.2 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 69 | d1d3va_ |
|
not modelled |
48.1 |
20 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 70 | c3fdgA_ |
|
not modelled |
45.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:dipeptidase ac. metallo peptidase. merops family m19;
PDBTitle: the crystal structure of the dipeptidase ac, metallo peptidase. merops2 family m19
|
| 71 | c3ez4B_ |
|
not modelled |
44.6 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate2 hydroxymethyltransferase from burkholderia pseudomallei
|
| 72 | d1f16a_ |
|
not modelled |
43.7 |
15 |
Fold:Toxins' membrane translocation domains
Superfamily:Bcl-2 inhibitors of programmed cell death
Family:Bcl-2 inhibitors of programmed cell death |
| 73 | d1m6ja_ |
|
not modelled |
43.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 74 | c3ai9X_ |
|
not modelled |
40.7 |
17 |
PDB header:transferase
Chain: X: PDB Molecule:upf0217 protein mj1640;
PDBTitle: crystal structure of duf358 protein reveals a putative spout-class2 rrna methyltransferase
|
| 75 | c3gvgA_ |
|
not modelled |
40.3 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from mycobacterium2 tuberculosis
|
| 76 | c2vzkD_ |
|
not modelled |
40.1 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:glutamate n-acetyltransferase 2 beta chain;
PDBTitle: structure of the acyl-enzyme complex of an n-terminal2 nucleophile (ntn) hydrolase, oat2
|
| 77 | c3b8iF_ |
|
not modelled |
39.9 |
18 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
| 78 | c3bleA_ |
|
not modelled |
39.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in2 complexed with malonate
|
| 79 | c2h31A_ |
|
not modelled |
38.5 |
17 |
PDB header:ligase, lyase
Chain: A: PDB Molecule:multifunctional protein ade2;
PDBTitle: crystal structure of human paics, a bifunctional carboxylase and2 synthetase in purine biosynthesis
|
| 80 | d2p02a1 |
|
not modelled |
38.5 |
16 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 81 | d1trea_ |
|
not modelled |
37.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 82 | c3tr9A_ |
|
not modelled |
37.6 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 83 | c1zorB_ |
|
not modelled |
36.6 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile thermotoga maritima
|
| 84 | c2gq0B_ |
|
not modelled |
36.3 |
16 |
PDB header:chaperone, hydrolase
Chain: B: PDB Molecule:chaperone protein htpg;
PDBTitle: crystal structure of the middle domain of htpg, the e. coli2 hsp90
|
| 85 | c3gndC_ |
|
not modelled |
36.0 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:aldolase lsrf;
PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
|
| 86 | c1k87A_ |
|
not modelled |
35.5 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:proline dehydrogenase;
PDBTitle: crystal structure of e.coli puta (residues 1-669)
|
| 87 | c1y6zA_ |
|
not modelled |
35.2 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:heat shock protein, putative;
PDBTitle: middle domain of plasmodium falciparum putative heat shock protein2 pf14_0417
|
| 88 | c2h9aB_ |
|
not modelled |
33.8 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase, iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 89 | c2o1tB_ |
|
not modelled |
33.5 |
16 |
PDB header:chaperone
Chain: B: PDB Molecule:endoplasmin;
PDBTitle: structure of middle plus c-terminal domains (m+c) of grp94
|
| 90 | d1mxaa1 |
|
not modelled |
33.1 |
14 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 91 | c2v1dA_ |
|
not modelled |
32.0 |
15 |
PDB header:oxidoreductase/repressor
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: structural basis of lsd1-corest selectivity in histone h32 recognition
|
| 92 | c3h7uA_ |
|
not modelled |
31.8 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldo-keto reductase;
PDBTitle: crystal structure of the plant stress-response enzyme akr4c9
|
| 93 | d1qm4a1 |
|
not modelled |
31.3 |
18 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 94 | d1xima_ |
|
not modelled |
30.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 95 | d2al1a1 |
|
not modelled |
30.3 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 96 | d2af4c1 |
|
not modelled |
30.2 |
12 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 97 | d1vz6a_ |
|
not modelled |
30.1 |
17 |
Fold:DmpA/ArgJ-like
Superfamily:DmpA/ArgJ-like
Family:ArgJ-like |
| 98 | c4a1aH_ |
|
not modelled |
30.0 |
24 |
PDB header:ribosome
Chain: H: PDB Molecule:60s ribosomal protein l10;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
|
| 99 | c3qc3B_ |
|
not modelled |
29.8 |
28 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
| 100 | d1t9ba3 |
|
not modelled |
29.7 |
19 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
| 101 | c1yl7F_ |
|
not modelled |
29.5 |
24 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: the crystal structure of mycobacterium tuberculosis2 dihydrodipicolinate reductase (rv2773c) in complex with nadh (crystal3 form c)
|
| 102 | d1n8fa_ |
|
not modelled |
29.4 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 103 | d3clsc1 |
|
not modelled |
28.3 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ETFP subunits |
| 104 | c1vm6B_ |
|
not modelled |
28.2 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: crystal structure of dihydrodipicolinate reductase (tm1520) from2 thermotoga maritima at 2.27 a resolution
|
| 105 | c3ih1A_ |
|
not modelled |
28.1 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 106 | d1s2wa_ |
|
not modelled |
28.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 107 | d1ixka_ |
|
not modelled |
27.4 |
29 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:NOL1/NOP2/sun |
| 108 | c1nofA_ |
|
not modelled |
27.2 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:xylanase;
PDBTitle: the first crystallographic structure of a xylanase from2 glycosyl hydrolase family 5: implications for catalysis
|
| 109 | c3m4xA_ |
|
not modelled |
26.9 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:nol1/nop2/sun family protein;
PDBTitle: structure of a ribosomal methyltransferase
|
| 110 | c3v7nA_ |
|
not modelled |
26.4 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:threonine synthase;
PDBTitle: crystal structure of threonine synthase (thrc) from from burkholderia2 thailandensis
|
| 111 | c2qb5B_ |
|
not modelled |
25.8 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:inositol-tetrakisphosphate 1-kinase;
PDBTitle: crystal structure of human inositol 1,3,4-trisphosphate 5/6-kinase2 (itpk1) in complex with adp and mn2+
|
| 112 | d1e0ta2 |
|
not modelled |
25.6 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 113 | d1oy0a_ |
|
not modelled |
25.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 114 | d2fyma1 |
|
not modelled |
25.3 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 115 | c3so4C_ |
|
not modelled |
25.2 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:methionine-adenosyltransferase;
PDBTitle: methionine-adenosyltransferase from entamoeba histolytica
|
| 116 | c1rg9D_ |
|
not modelled |
24.8 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:s-adenosylmethionine synthetase;
PDBTitle: s-adenosylmethionine synthetase complexed with sam and ppnp
|
| 117 | d1azta1 |
|
not modelled |
24.7 |
15 |
Fold:Transducin (alpha subunit), insertion domain
Superfamily:Transducin (alpha subunit), insertion domain
Family:Transducin (alpha subunit), insertion domain |
| 118 | d2ebna_ |
|
not modelled |
24.6 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Type II chitinase |
| 119 | c2e1mC_ |
|
not modelled |
24.1 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:l-glutamate oxidase;
PDBTitle: crystal structure of l-glutamate oxidase from streptomyces sp. x-119-6
|
| 120 | d1vm6a3 |
|
not modelled |
24.0 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |