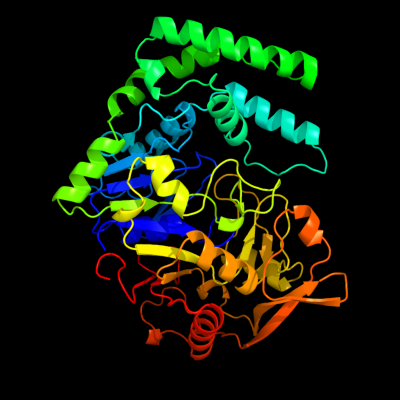
| 1 | d1qf5a_
|
|
 |
100.0 |
100 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 2 | d1dj2a_
|
|
 |
100.0 |
44 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 3 | d1dj3a_
|
|
 |
100.0 |
44 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 4 | d1iwea_
|
|
 |
100.0 |
43 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 5 | c1iweB_
|
|
 |
100.0 |
43 |
PDB header:ligase
Chain: B: PDB Molecule:adenylosuccinate synthetase;
PDBTitle: imp complex of the recombinant mouse-muscle2 adenylosuccinate synthetase
|
| 6 | d1p9ba_
|
|
 |
100.0 |
40 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 7 | c3ue9A_
|
|
 |
100.0 |
61 |
PDB header:ligase
Chain: A: PDB Molecule:adenylosuccinate synthetase;
PDBTitle: crystal structure of adenylosuccinate synthetase (ampsase) (pura) from2 burkholderia thailandensis
|
| 8 | c3r7tA_
|
|
 |
100.0 |
47 |
PDB header:ligase
Chain: A: PDB Molecule:adenylosuccinate synthetase;
PDBTitle: crystal structure of adenylosuccinate synthetase from campylobacter2 jejuni
|
| 9 | c2d7uA_
|
|
 |
100.0 |
43 |
PDB header:ligase
Chain: A: PDB Molecule:adenylosuccinate synthetase;
PDBTitle: crystal structure of hypothetical adenylosuccinate synthetase, ph04382 from pyrococcus horikoshii ot3
|
| 10 | d1a0ia1
|
|
 |
88.4 |
24 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:DNA ligase/mRNA capping enzyme postcatalytic domain |
| 11 | d1p9pa_
|
|
 |
57.7 |
19 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:tRNA(m1G37)-methyltransferase TrmD |
| 12 | c3fwzA_
|
|
 |
54.0 |
27 |
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein ybal;
PDBTitle: crystal structure of trka-n domain of inner membrane protein ybal from2 escherichia coli
|
| 13 | d1uala_
|
|
 |
54.0 |
19 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:tRNA(m1G37)-methyltransferase TrmD |
| 14 | d2jfga1
|
|
 |
53.2 |
21 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
| 15 | c3knuD_
|
|
 |
52.3 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: crystal structure of trna (guanine-n1)-methyltransferase from2 anaplasma phagocytophilum
|
| 16 | c1zgxA_
|
|
 |
49.6 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:guanyl-specific ribonuclease sa;
PDBTitle: crystal structure of ribonuclease mutant
|
| 17 | d2f09a1
|
|
 |
44.9 |
10 |
Fold:Streptavidin-like
Superfamily:YdhA-like
Family:YdhA-like |
| 18 | c3ky7A_
|
|
 |
43.7 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: 2.35 angstrom resolution crystal structure of a putative trna2 (guanine-7-)-methyltransferase (trmd) from staphylococcus aureus3 subsp. aureus mrsa252
|
| 19 | d1qwda_
|
|
 |
39.7 |
15 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Retinol binding protein-like |
| 20 | d1gm5a2
|
|
 |
35.5 |
28 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:RecG "wedge" domain |
| 21 | c3iefA_ |
|
not modelled |
34.3 |
25 |
PDB header:transferase, rna binding protein
Chain: A: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: crystal structure of trna guanine-n1-methyltransferase from2 bartonella henselae using mpcs.
|
| 22 | c2ixaA_ |
|
not modelled |
34.2 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylgalactosaminidase;
PDBTitle: a-zyme, n-acetylgalactosaminidase
|
| 23 | c2qioA_ |
|
not modelled |
32.3 |
8 |
PDB header:unknown function
Chain: A: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: x-ray structure of enoyl-acyl carrier protein reductase from bacillus2 anthracis with triclosan
|
| 24 | d1jsca3 |
|
not modelled |
30.9 |
14 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
| 25 | c1k1qA_ |
|
not modelled |
28.4 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:dbh protein;
PDBTitle: crystal structure of a dinb family error prone dna2 polymerase from sulfolobus solfataricus
|
| 26 | c3dnfB_ |
|
not modelled |
26.3 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate reductase;
PDBTitle: structure of (e)-4-hydroxy-3-methyl-but-2-enyl diphosphate reductase,2 the terminal enzyme of the non-mevalonate pathway
|
| 27 | c3quvB_ |
|
not modelled |
26.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: crystal structure of a trna-guanine-n1-methyltransferase from2 mycobacterium abscessus
|
| 28 | d1lt7a_ |
|
not modelled |
24.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 29 | c2xf4A_ |
|
not modelled |
24.6 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydroxyacylglutathione hydrolase;
PDBTitle: crystal structure of salmonella enterica serovar2 typhimurium ycbl
|
| 30 | c3opyE_ |
|
not modelled |
24.3 |
12 |
PDB header:transferase
Chain: E: PDB Molecule:6-phosphofructo-1-kinase alpha-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
| 31 | d1k1ga_ |
|
not modelled |
23.4 |
29 |
Fold:Eukaryotic type KH-domain (KH-domain type I)
Superfamily:Eukaryotic type KH-domain (KH-domain type I)
Family:Eukaryotic type KH-domain (KH-domain type I) |
| 32 | d1g7oa1 |
|
not modelled |
22.3 |
14 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 33 | d1e6ca_ |
|
not modelled |
22.3 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Shikimate kinase (AroK) |
| 34 | c2nm0B_ |
|
not modelled |
21.6 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable 3-oxacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of sco1815: a beta-ketoacyl-acyl carrier protein2 reductase from streptomyces coelicolor a3(2)
|
| 35 | c1kyqC_ |
|
not modelled |
21.5 |
17 |
PDB header:oxidoreductase, lyase
Chain: C: PDB Molecule:siroheme biosynthesis protein met8;
PDBTitle: met8p: a bifunctional nad-dependent dehydrogenase and2 ferrochelatase involved in siroheme synthesis.
|
| 36 | d1hywa_ |
|
not modelled |
20.0 |
41 |
Fold:gpW/XkdW-like
Superfamily:Head-to-tail joining protein W, gpW
Family:Head-to-tail joining protein W, gpW |
| 37 | c2inrA_ |
|
not modelled |
19.6 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:dna topoisomerase 4 subunit a;
PDBTitle: crystal structure of a 59 kda fragment of topoisomerase iv subunit a2 (grla) from staphylococcus aureus
|
| 38 | d1lnia_ |
|
not modelled |
18.8 |
25 |
Fold:Microbial ribonucleases
Superfamily:Microbial ribonucleases
Family:Bacterial ribonucleases |
| 39 | c3p19A_ |
|
not modelled |
18.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative blue fluorescent protein;
PDBTitle: improved nadph-dependent blue fluorescent protein
|
| 40 | c2z2vA_ |
|
not modelled |
18.6 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein ph1688;
PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
|
| 41 | c2cfcB_ |
|
not modelled |
18.3 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-(r)-hydroxypropyl-com dehydrogenase;
PDBTitle: structural basis for stereo selectivity in the (r)- and2 (s)-hydroxypropylethane thiosulfonate dehydrogenases
|
| 42 | c3db2C_ |
|
not modelled |
18.0 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
| 43 | c2gr2A_ |
|
not modelled |
18.0 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin reductase;
PDBTitle: crystal structure of ferredoxin reductase, bpha4 (oxidized form)
|
| 44 | c2wfbA_ |
|
not modelled |
17.7 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative uncharacterized protein orp;
PDBTitle: high resolution structure of the apo form of the orange2 protein (orp) from desulfovibrio gigas
|
| 45 | d1d7ya1 |
|
not modelled |
17.7 |
42 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 46 | d1oy5a_ |
|
not modelled |
17.6 |
14 |
Fold:alpha/beta knot
Superfamily:alpha/beta knot
Family:tRNA(m1G37)-methyltransferase TrmD |
| 47 | c1oy5B_ |
|
not modelled |
17.6 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:trna (guanine-n(1)-)-methyltransferase;
PDBTitle: crystal structure of trna (m1g37) methyltransferase from aquifex2 aeolicus
|
| 48 | c1yfsB_ |
|
not modelled |
17.5 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:alanyl-trna synthetase;
PDBTitle: the crystal structure of alanyl-trna synthetase in complex2 with l-alanine
|
| 49 | d2i1qa1 |
|
not modelled |
17.4 |
15 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:DNA repair protein Rad51, N-terminal domain |
| 50 | c3ktsA_ |
|
not modelled |
17.4 |
14 |
PDB header:transcriptional regulator
Chain: A: PDB Molecule:glycerol uptake operon antiterminator regulatory protein;
PDBTitle: crystal structure of glycerol uptake operon antiterminator regulatory2 protein from listeria monocytogenes str. 4b f2365
|
| 51 | d1k1sa2 |
|
not modelled |
17.0 |
16 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 52 | d1qyia_ |
|
not modelled |
16.9 |
22 |
Fold:HAD-like
Superfamily:HAD-like
Family:Hypothetical protein MW1667 (SA1546) |
| 53 | d1qnaa1 |
|
not modelled |
16.7 |
12 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
| 54 | d1nh2a1 |
|
not modelled |
16.5 |
11 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
| 55 | d1jx4a2 |
|
not modelled |
16.3 |
18 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 56 | c2zzfA_ |
|
not modelled |
16.0 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:alanyl-trna synthetase;
PDBTitle: crystal structure of alanyl-trna synthetase without2 oligomerization domain
|
| 57 | c3f6zB_ |
|
not modelled |
15.9 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of pseudomonas aeruginosa mlic in complex2 with hen egg white lysozyme
|
| 58 | d1q44a_ |
|
not modelled |
15.8 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:PAPS sulfotransferase |
| 59 | c1ujlA_ |
|
not modelled |
15.6 |
33 |
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h
PDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
|
| 60 | c3dtyA_ |
|
not modelled |
15.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from pseudomonas2 syringae
|
| 61 | d1fi4a2 |
|
not modelled |
15.1 |
56 |
Fold:Ferredoxin-like
Superfamily:GHMP Kinase, C-terminal domain
Family:Mevalonate 5-diphosphate decarboxylase |
| 62 | d1y6ia1 |
|
not modelled |
15.0 |
78 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:GUN4-associated domain |
| 63 | d1cora_ |
|
not modelled |
14.8 |
18 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:monodomain cytochrome c |
| 64 | d2bl5a1 |
|
not modelled |
14.7 |
29 |
Fold:Eukaryotic type KH-domain (KH-domain type I)
Superfamily:Eukaryotic type KH-domain (KH-domain type I)
Family:Eukaryotic type KH-domain (KH-domain type I) |
| 65 | d1cdwa1 |
|
not modelled |
14.6 |
12 |
Fold:TBP-like
Superfamily:TATA-box binding protein-like
Family:TATA-box binding protein (TBP), C-terminal domain |
| 66 | d1uuqa_ |
|
not modelled |
14.0 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 67 | c1uz4A_ |
|
not modelled |
14.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:man5a;
PDBTitle: common inhibition of beta-glucosidase and beta-mannosidase2 by isofagomine lactam reflects different conformational3 intineraries for glucoside and mannoside hydrolysis
|
| 68 | d1pzna1 |
|
not modelled |
13.9 |
21 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:DNA repair protein Rad51, N-terminal domain |
| 69 | c2hivA_ |
|
not modelled |
13.6 |
42 |
PDB header:ligase
Chain: A: PDB Molecule:thermostable dna ligase;
PDBTitle: atp-dependent dna ligase from s. solfataricus
|
| 70 | d2bcqa2 |
|
not modelled |
13.6 |
25 |
Fold:SAM domain-like
Superfamily:PsbU/PolX domain-like
Family:DNA polymerase beta-like, second domain |
| 71 | c2v7sA_ |
|
not modelled |
13.5 |
28 |
PDB header:unknown function
Chain: A: PDB Molecule:probable conserved lipoprotein lppa;
PDBTitle: crystal structure of the putative lipoprotein lppa from2 mycobacterium tuberculosis
|
| 72 | c1t3nB_ |
|
not modelled |
13.4 |
22 |
PDB header:replication/dna
Chain: B: PDB Molecule:polymerase (dna directed) iota;
PDBTitle: structure of the catalytic core of dna polymerase iota in2 complex with dna and dttp
|
| 73 | d1t6ca1 |
|
not modelled |
13.4 |
18 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Ppx/GppA phosphatase |
| 74 | d1id1a_ |
|
not modelled |
13.2 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
| 75 | d1jmsa3 |
|
not modelled |
13.1 |
38 |
Fold:SAM domain-like
Superfamily:PsbU/PolX domain-like
Family:DNA polymerase beta-like, second domain |
| 76 | d1znwa1 |
|
not modelled |
12.8 |
32 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 77 | c3c5yD_ |
|
not modelled |
12.6 |
18 |
PDB header:isomerase
Chain: D: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
|
| 78 | d1qsga_ |
|
not modelled |
12.6 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 79 | c2y0nD_ |
|
not modelled |
12.6 |
13 |
PDB header:transcription
Chain: D: PDB Molecule:male-specific lethal 3 homolog;
PDBTitle: crystal structure of the complex between dosage2 compensation factors msl1 and msl3
|
| 80 | d1riqa1 |
|
not modelled |
12.4 |
13 |
Fold:Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)
Superfamily:Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)
Family:Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS) |
| 81 | c3hi0B_ |
|
not modelled |
12.3 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative exopolyphosphatase;
PDBTitle: crystal structure of putative exopolyphosphatase (17739545) from2 agrobacterium tumefaciens str. c58 (dupont) at 2.30 a resolution
|
| 82 | d1zeta2 |
|
not modelled |
12.1 |
22 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 83 | d1t94a2 |
|
not modelled |
12.0 |
14 |
Fold:DNA/RNA polymerases
Superfamily:DNA/RNA polymerases
Family:Lesion bypass DNA polymerase (Y-family), catalytic domain |
| 84 | d1x9na2 |
|
not modelled |
11.9 |
32 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:DNA ligase/mRNA capping enzyme postcatalytic domain |
| 85 | d2a3ra1 |
|
not modelled |
11.9 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:PAPS sulfotransferase |
| 86 | c1u9iA_ |
|
not modelled |
11.6 |
12 |
PDB header:circadian clock protein
Chain: A: PDB Molecule:kaic;
PDBTitle: crystal structure of circadian clock protein kaic with phosphorylation2 sites
|
| 87 | c1s97D_ |
|
not modelled |
11.5 |
14 |
PDB header:transferase/dna
Chain: D: PDB Molecule:dna polymerase iv;
PDBTitle: dpo4 with gt mismatch
|
| 88 | c2ovfA_ |
|
not modelled |
11.4 |
4 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:stal;
PDBTitle: crystal structure of stal-pap complex
|
| 89 | d1a0rp_ |
|
not modelled |
11.4 |
33 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Phosducin |
| 90 | c3o38D_ |
|
not modelled |
11.2 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase from mycobacterium2 smegmatis
|
| 91 | c1ylmA_ |
|
not modelled |
11.1 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein bsu32300;
PDBTitle: structure of cytosolic protein of unknown function yute2 from bacillus subtilis
|
| 92 | d2fmpa2 |
|
not modelled |
11.0 |
38 |
Fold:SAM domain-like
Superfamily:PsbU/PolX domain-like
Family:DNA polymerase beta-like, second domain |
| 93 | d1fuea_ |
|
not modelled |
10.9 |
13 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:Flavodoxin-related |
| 94 | d2vana1 |
|
not modelled |
10.9 |
38 |
Fold:SAM domain-like
Superfamily:PsbU/PolX domain-like
Family:DNA polymerase beta-like, second domain |
| 95 | d2ppqa1 |
|
not modelled |
10.8 |
14 |
Fold:Protein kinase-like (PK-like)
Superfamily:Protein kinase-like (PK-like)
Family:APH phosphotransferases |
| 96 | c3he8A_ |
|
not modelled |
10.5 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
|
| 97 | c1fi4A_ |
|
not modelled |
10.5 |
56 |
PDB header:lyase
Chain: A: PDB Molecule:mevalonate 5-diphosphate decarboxylase;
PDBTitle: the x-ray crystal structure of mevalonate 5-diphosphate decarboxylase2 at 2.3 angstrom resolution.
|
| 98 | c3nbxX_ |
|
not modelled |
10.5 |
21 |
PDB header:hydrolase
Chain: X: PDB Molecule:atpase rava;
PDBTitle: crystal structure of e. coli rava (regulatory atpase variant a) in2 complex with adp
|
| 99 | c2fllA_ |
|
not modelled |
10.5 |
22 |
PDB header:replication/dna
Chain: A: PDB Molecule:dna polymerase iota;
PDBTitle: ternary complex of human dna polymerase iota with dna and dttp
|