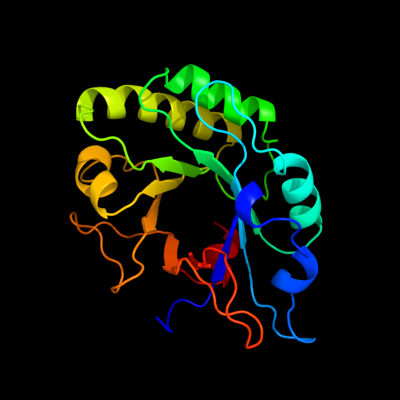
| 1 | c2wagA_
|
|
 |
100.0 |
35 |
PDB header:hydrolase
Chain: A: PDB Molecule:lysozyme, putative;
PDBTitle: the structure of a family 25 glycosyl hydrolase from2 bacillus anthracis.
|
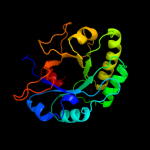
| 2 | d1jfxa_
|
|
 |
100.0 |
36 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:1,4-beta-N-acetylmuraminidase |
| 3 | c2x8rE_
|
|
 |
100.0 |
31 |
PDB header:hydrolase
Chain: E: PDB Molecule:glycosyl hydrolase;
PDBTitle: the structure of a family gh25 lysozyme from aspergillus2 fumigatus
|
| 4 | c2wwcA_
|
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:1,4-beta-n-acetylmuramidase;
PDBTitle: 3d-structure of the modular autolysin lytc from2 streptococcus pneumoniae in complex with synthetic3 peptidoglycan ligand
|
| 5 | c2nw0B_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:plyb;
PDBTitle: crystal structure of a lysin
|
| 6 | d2j8ga2
|
|
 |
100.0 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:1,4-beta-N-acetylmuraminidase |
| 7 | c2j8fA_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:lysozyme;
PDBTitle: crystal structure of the modular cpl-1 endolysin complexed2 with a peptidoglycan analogue (e94q mutant in complex with3 a disaccharide-pentapeptide)
|
| 8 | d1sfsa_
|
|
 |
98.6 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:1,4-beta-N-acetylmuraminidase |
| 9 | c1sfsA_
|
|
 |
98.6 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: 1.07 a crystal structure of an uncharacterized b.2 stearothermophilus protein
|
| 10 | d1hl9a2
|
|
 |
64.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Putative alpha-L-fucosidase, catalytic domain |
| 11 | d1e4ia_
|
|
 |
63.8 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 12 | d1hjqa_
|
|
 |
62.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 13 | d1hjsa_
|
|
 |
62.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 14 | c2wvsD_
|
|
 |
57.5 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:alpha-l-fucosidase;
PDBTitle: crystal structure of an alpha-l-fucosidase gh29 trapped2 covalent intermediate from bacteroides thetaiotaomicron in3 complex with 2-fluoro-fucosyl fluoride using an e288q4 mutant
|
| 15 | c3s6dA_
|
|
 |
56.2 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:putative triosephosphate isomerase;
PDBTitle: crystal structure of a putative triosephosphate isomerase from2 coccidioides immitis
|
| 16 | c3gzaB_
|
|
 |
54.8 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of putative alpha-l-fucosidase (np_812709.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.60 a resolution
|
| 17 | c3ahxC_
|
|
 |
53.4 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-glucosidase a;
PDBTitle: crystal structure of beta-glucosidase a from bacterium clostridium2 cellulovorans
|
| 18 | c1hl8B_
|
|
 |
51.9 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of thermotoga maritima alpha-fucosidase
|
| 19 | d1euca2
|
|
 |
50.5 |
17 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 20 | d2j78a1
|
|
 |
44.6 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 21 | d1tvna1 |
|
not modelled |
44.4 |
6 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 22 | c3eypB_ |
|
not modelled |
42.2 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of putative alpha-l-fucosidase from bacteroides2 thetaiotaomicron
|
| 23 | c3ebvA_ |
|
not modelled |
39.0 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:chinitase a;
PDBTitle: crystal structure of putative chitinase a from streptomyces2 coelicolor.
|
| 24 | c2jgqB_ |
|
not modelled |
37.5 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: kinetics and structural properties of triosephosphate2 isomerase from helicobacter pylori
|
| 25 | d1egza_ |
|
not modelled |
37.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 26 | d1foba_ |
|
not modelled |
36.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 27 | c3mo4B_ |
|
not modelled |
34.6 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-1,3/4-fucosidase;
PDBTitle: the crystal structure of an alpha-(1-3,4)-fucosidase from2 bifidobacterium longum subsp. infantis atcc 15697
|
| 28 | c3th6B_ |
|
not modelled |
34.5 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from rhipicephalus2 (boophilus) microplus.
|
| 29 | c3qr3B_ |
|
not modelled |
29.2 |
8 |
PDB header:hydrolase
Chain: B: PDB Molecule:endoglucanase eg-ii;
PDBTitle: crystal structure of cel5a (eg2) from hypocrea jecorina (trichoderma2 reesei)
|
| 30 | d1o5xa_ |
|
not modelled |
28.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 31 | c3krsB_ |
|
not modelled |
28.2 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: structure of triosephosphate isomerase from cryptosporidium parvum at2 1.55a resolution
|
| 32 | d1ur4a_ |
|
not modelled |
27.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 33 | c3ianA_ |
|
not modelled |
25.4 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitinase;
PDBTitle: crystal structure of a chitinase from lactococcus lactis2 subsp. lactis
|
| 34 | d1wkya2 |
|
not modelled |
21.0 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 35 | c2xucA_ |
|
not modelled |
20.4 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitinase;
PDBTitle: natural product-guided discovery of a fungal chitinase2 inhibitor
|
| 36 | c3c65A_ |
|
not modelled |
20.3 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:uvrabc system protein c;
PDBTitle: crystal structure of bacillus stearothermophilus uvrc 5'2 endonuclease domain
|
| 37 | d1gnxa_ |
|
not modelled |
19.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 38 | d1f61a_ |
|
not modelled |
17.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 39 | c3e5bB_ |
|
not modelled |
16.9 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:isocitrate lyase;
PDBTitle: 2.4 a crystal structure of isocitrate lyase from brucella2 melitensis
|
| 40 | d1ktba2 |
|
not modelled |
16.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 41 | c2g3nA_ |
|
not modelled |
15.6 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-glucosidase;
PDBTitle: crystal structure of the sulfolobus solfataricus alpha-2 glucosidase mala in complex with beta-octyl-glucopyranoside
|
| 42 | c2pt7G_ |
|
not modelled |
15.3 |
12 |
PDB header:hydrolase/protein binding
Chain: G: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of cag virb11 (hp0525) and an inhibitory protein2 (hp1451)
|
| 43 | d1b9ba_ |
|
not modelled |
13.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 44 | c3mmwB_ |
|
not modelled |
13.3 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:endoglucanase;
PDBTitle: crystal structure of endoglucanase cel5a from the hyperthermophilic2 thermotoga maritima
|
| 45 | d1q1ra3 |
|
not modelled |
13.3 |
22 |
Fold:CO dehydrogenase flavoprotein C-domain-like
Superfamily:FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Family:FAD/NAD-linked reductases, dimerisation (C-terminal) domain |
| 46 | c2dskA_ |
|
not modelled |
13.1 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitinase;
PDBTitle: crystal structure of catalytic domain of hyperthermophilic chitinase2 from pyrococcus furiosus
|
| 47 | d1qcrd2 |
|
not modelled |
13.0 |
22 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Cytochrome bc1 domain |
| 48 | d1mo0a_ |
|
not modelled |
12.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 49 | c3ffjA_ |
|
not modelled |
12.2 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyl hydrolase (family 31);
PDBTitle: the crystal structure of the glycosyl hydrolase (family 31) from2 ruminococcus obeum atcc 29174
|
| 50 | c2qmjA_ |
|
not modelled |
12.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:maltase-glucoamylase, intestinal;
PDBTitle: crystral structure of the n-terminal subunit of human maltase-2 glucoamylase in complex with acarbose
|
| 51 | c3lecA_ |
|
not modelled |
11.9 |
18 |
PDB header:structure genomics, unknown function
Chain: A: PDB Molecule:nadb-rossmann superfamily protein;
PDBTitle: the crystal structure of a protein in the nadb-rossmann2 superfamily from streptococcus agalactiae to 1.8a
|
| 52 | d1trea_ |
|
not modelled |
11.6 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 53 | d1o0sa2 |
|
not modelled |
11.6 |
28 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Malic enzyme N-domain |
| 54 | c1qcrD_ |
|
not modelled |
11.5 |
21 |
PDB header:
PDB COMPND:
|
| 55 | d1suxa_ |
|
not modelled |
11.2 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 56 | c2rirA_ |
|
not modelled |
10.9 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
| 57 | c3lppA_ |
|
not modelled |
10.8 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:sucrase-isomaltase;
PDBTitle: crystal complex of n-terminal sucrase-isomaltase with kotalanol
|
| 58 | d1qoxa_ |
|
not modelled |
10.8 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 59 | d1aw1a_ |
|
not modelled |
10.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 60 | c3gnoA_ |
|
not modelled |
10.6 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:os03g0212800 protein;
PDBTitle: crystal structure of a rice os3bglu6 beta-glucosidase
|
| 61 | d1nqua_ |
|
not modelled |
10.2 |
14 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
| 62 | d1pj3a2 |
|
not modelled |
10.2 |
18 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Malic enzyme N-domain |
| 63 | d1neya_ |
|
not modelled |
10.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 64 | c2dp3A_ |
|
not modelled |
10.0 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of a double mutant (c202a/a198v) of triosephosphate2 isomerase from giardia lamblia
|
| 65 | d1eg7a_ |
|
not modelled |
9.9 |
13 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 66 | d1kv5a_ |
|
not modelled |
9.7 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 67 | c2rgmA_ |
|
not modelled |
9.5 |
34 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: rice bglu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase
|
| 68 | c3kxqB_ |
|
not modelled |
9.4 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from bartonella2 henselae at 1.6a resolution
|
| 69 | d2btma_ |
|
not modelled |
9.3 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 70 | c3civA_ |
|
not modelled |
9.2 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-beta-1,4-mannanase;
PDBTitle: crystal structure of the endo-beta-1,4-mannanase from2 alicyclobacillus acidocaldarius
|
| 71 | c2h6rG_ |
|
not modelled |
8.9 |
15 |
PDB header:isomerase
Chain: G: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase (tim) from2 methanocaldococcus jannaschii
|
| 72 | c2j75A_ |
|
not modelled |
8.8 |
34 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase a;
PDBTitle: beta-glucosidase from thermotoga maritima in complex with2 noeuromycin
|
| 73 | d2ebna_ |
|
not modelled |
8.8 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Type II chitinase |
| 74 | c2r94B_ |
|
not modelled |
8.6 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
| 75 | c1yyaA_ |
|
not modelled |
8.5 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of tt0473, putative triosephosphate isomerase from2 thermus thermophilus hb8
|
| 76 | c3bghB_ |
|
not modelled |
8.5 |
35 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative neuraminyllactose-binding hemagglutinin homolog;
PDBTitle: crystal structure of putative neuraminyllactose-binding hemagglutinin2 homolog from helicobacter pylori
|
| 77 | c3d4oA_ |
|
not modelled |
8.1 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 78 | c1zrtD_ |
|
not modelled |
8.1 |
19 |
PDB header:oxidoreductase/metal transport
Chain: D: PDB Molecule:cytochrome c1;
PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
|
| 79 | c3jugA_ |
|
not modelled |
8.1 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-mannanase;
PDBTitle: crystal structure of endo-beta-1,4-mannanase from the alkaliphilic2 bacillus sp. n16-5
|
| 80 | c2nrzB_ |
|
not modelled |
8.0 |
50 |
PDB header:hydrolase
Chain: B: PDB Molecule:uvrabc system protein c;
PDBTitle: crystal structure of the c-terminal half of uvrc bound to2 its catalytic divalent cation
|
| 81 | c3cz8A_ |
|
not modelled |
7.9 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative sporulation-specific glycosylase ydhd;
PDBTitle: crystal structure of putative sporulation-specific glycosylase ydhd2 from bacillus subtilis
|
| 82 | c2dgaA_ |
|
not modelled |
7.6 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of hexameric beta-glucosidase in wheat
|
| 83 | d1oa3a_ |
|
not modelled |
7.5 |
19 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Xylanase/endoglucanase 11/12 |
| 84 | c3d5hA_ |
|
not modelled |
7.2 |
14 |
PDB header:protein binding
Chain: A: PDB Molecule:haementhin;
PDBTitle: crystal structure of haementhin from haemanthus multiflorus2 at 2.0a resolution: formation of a novel loop on a tim3 barrel fold and its functional significance
|
| 85 | d1pbga_ |
|
not modelled |
7.2 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 86 | d2i9ia1 |
|
not modelled |
7.1 |
19 |
Fold:Anticodon-binding domain-like
Superfamily:XCC0632-like
Family:NLBH-like |
| 87 | c2i9iA_ |
|
not modelled |
7.1 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of helicobacter pylori protein hp0492
|
| 88 | c1wkyA_ |
|
not modelled |
7.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:endo-beta-1,4-mannanase;
PDBTitle: crystal structure of alkaline mannanase from bacillus sp. strain jamb-2 602: catalytic domain and its carbohydrate binding module
|
| 89 | d1u83a_ |
|
not modelled |
7.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:(2r)-phospho-3-sulfolactate synthase ComA
Family:(2r)-phospho-3-sulfolactate synthase ComA |
| 90 | c1u83A_ |
|
not modelled |
7.1 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:phosphosulfolactate synthase;
PDBTitle: psl synthase from bacillus subtilis
|
| 91 | d1ta3a_ |
|
not modelled |
7.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Type II chitinase |
| 92 | c2fpgA_ |
|
not modelled |
6.9 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa ligase [gdp-forming] alpha-chain,
PDBTitle: crystal structure of pig gtp-specific succinyl-coa2 synthetase in complex with gdp
|
| 93 | d1edta_ |
|
not modelled |
6.9 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Type II chitinase |
| 94 | d1rvv1_ |
|
not modelled |
6.7 |
14 |
Fold:Lumazine synthase
Superfamily:Lumazine synthase
Family:Lumazine synthase |
| 95 | d1r2ra_ |
|
not modelled |
6.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 96 | c2uy2A_ |
|
not modelled |
6.6 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:endochitinase;
PDBTitle: sccts1_apo crystal structure
|
| 97 | d1m6ja_ |
|
not modelled |
6.6 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 98 | c3thdD_ |
|
not modelled |
6.6 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-galactosidase;
PDBTitle: crystal structure of human beta-galactosidase in complex with 1-2 deoxygalactonojirimycin
|
| 99 | c3topA_ |
|
not modelled |
6.6 |
6 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:maltase-glucoamylase, intestinal;
PDBTitle: crystral structure of the c-terminal subunit of human maltase-2 glucoamylase in complex with acarbose
|