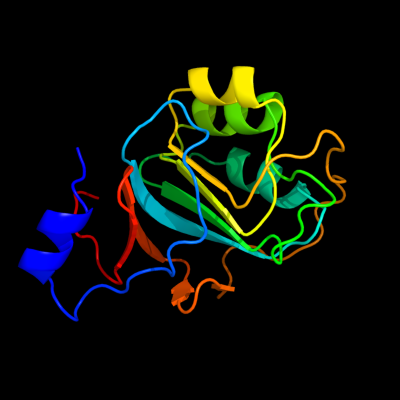
| 1 | d1q5xa_
|
|
 |
100.0 |
100 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
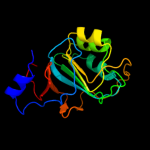
| 2 | c3c8oB_
|
|
 |
100.0 |
55 |
PDB header:hydrolase regulator
Chain: B: PDB Molecule:regulator of ribonuclease activity a;
PDBTitle: the crystal structure of rraa from pao1
|
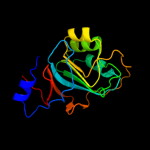
| 3 | c2pcnA_
|
|
 |
100.0 |
38 |
PDB header:transferase
Chain: A: PDB Molecule:s-adenosylmethionine:2-demethylmenaquinone
PDBTitle: crystal structure of s-adenosylmethionine: 2-dimethylmenaquinone2 methyltransferase (gk_1813) from geobacillus kaustophilus hta426
|
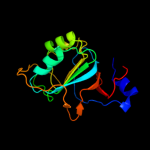
| 4 | d1vi4a_
|
|
 |
100.0 |
44 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
| 5 | d1j3la_
|
|
 |
100.0 |
44 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
| 6 | d1nxja_
|
|
 |
100.0 |
45 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:RraA-like
Family:RraA-like |
| 7 | c1nxjA_
|
|
 |
100.0 |
45 |
PDB header:unknown function
Chain: A: PDB Molecule:probable s-adenosylmethionine:2-
PDBTitle: structure of rv3853 from mycobacterium tuberculosis
|
| 8 | c3nojA_
|
|
 |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate
PDBTitle: the structure of hmg/cha aldolase from the protocatechuate degradation2 pathway of pseudomonas putida
|
| 9 | c3k4iC_
|
|
 |
100.0 |
19 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein pspto_3204 from2 pseudomonas syringae pv. tomato str. dc3000
|
| 10 | c2c5qE_
|
|
 |
100.0 |
19 |
PDB header:structural genomics,unknown function
Chain: E: PDB Molecule:rraa-like protein yer010c;
PDBTitle: crystal structure of yeast yer010cp
|
| 11 | d1vbga2
|
|
 |
76.9 |
15 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
| 12 | d1kbla2
|
|
 |
75.0 |
20 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
| 13 | d2gp4a1
|
|
 |
60.6 |
20 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:IlvD/EDD C-terminal domain-like |
| 14 | d1f5va_
|
|
 |
55.4 |
18 |
Fold:FMN-dependent nitroreductase-like
Superfamily:FMN-dependent nitroreductase-like
Family:NADH oxidase/flavin reductase |
| 15 | d1e8ca2
|
|
 |
55.2 |
20 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 16 | c3n2sD_
|
|
 |
53.4 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nadph-dependent nitro/flavin reductase;
PDBTitle: structure of nfra1 nitroreductase from b. subtilis
|
| 17 | d1zcha1
|
|
 |
49.8 |
15 |
Fold:FMN-dependent nitroreductase-like
Superfamily:FMN-dependent nitroreductase-like
Family:NADH oxidase/flavin reductase |
| 18 | c2gp4A_
|
|
 |
47.1 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:6-phosphogluconate dehydratase;
PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
|
| 19 | c3eofB_
|
|
 |
43.7 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase (yp_213212.1) from2 bacteroides fragilis nctc 9343 at 1.99 a resolution
|
| 20 | d1bkja_
|
|
 |
42.7 |
18 |
Fold:FMN-dependent nitroreductase-like
Superfamily:FMN-dependent nitroreductase-like
Family:NADH oxidase/flavin reductase |
| 21 | c1vfgB_ |
|
not modelled |
42.6 |
17 |
PDB header:transferase/rna
Chain: B: PDB Molecule:poly a polymerase;
PDBTitle: crystal structure of trna nucleotidyltransferase complexed2 with a primer trna and an incoming atp analog
|
| 22 | c1ou5A_ |
|
not modelled |
40.9 |
36 |
PDB header:translation, transferase
Chain: A: PDB Molecule:trna cca-adding enzyme;
PDBTitle: crystal structure of human cca-adding enzyme
|
| 23 | d1vfga2 |
|
not modelled |
37.7 |
17 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Poly A polymerase head domain-like |
| 24 | d1ou5a2 |
|
not modelled |
33.3 |
36 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Poly A polymerase head domain-like |
| 25 | c1dbgA_ |
|
not modelled |
33.1 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:chondroitinase b;
PDBTitle: crystal structure of chondroitinase b
|
| 26 | c2vpiA_ |
|
not modelled |
31.2 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:gmp synthase;
PDBTitle: human gmp synthetase - glutaminase domain
|
| 27 | d1ofla_ |
|
not modelled |
30.1 |
22 |
Fold:Single-stranded right-handed beta-helix
Superfamily:Pectin lyase-like
Family:Chondroitinase B |
| 28 | c1e8cB_ |
|
not modelled |
27.4 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:udp-n-acetylmuramoylalanyl-d-glutamate--2,6-
PDBTitle: structure of mure the udp-n-acetylmuramyl tripeptide2 synthetase from e. coli
|
| 29 | d1miwa2 |
|
not modelled |
24.8 |
23 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Poly A polymerase head domain-like |
| 30 | c3mvnA_ |
|
not modelled |
23.7 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamayl-medo-
PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamayl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
|
| 31 | c3h37B_ |
|
not modelled |
21.7 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:trna nucleotidyl transferase-related protein;
PDBTitle: the structure of cca-adding enzyme apo form i
|
| 32 | c3aqnA_ |
|
not modelled |
19.5 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:poly(a) polymerase;
PDBTitle: complex structure of bacterial protein (apo form ii)
|
| 33 | d1thfd_ |
|
not modelled |
18.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 34 | c3l7oB_ |
|
not modelled |
18.4 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
|
| 35 | c3k6hB_ |
|
not modelled |
18.1 |
5 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitroreductase family protein;
PDBTitle: crystal structure of a nitroreductase family protein from2 agrobacterium tumefaciens str. c58
|
| 36 | c3tbiB_ |
|
not modelled |
16.6 |
33 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: crystal structure of t4 gp33 bound to e. coli rnap beta-flap domain
|
| 37 | c2pjmA_ |
|
not modelled |
16.4 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: structure of ribose 5-phosphate isomerase a from2 methanocaldococcus jannaschii
|
| 38 | c3bm2B_ |
|
not modelled |
16.4 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:protein ydja;
PDBTitle: crystal structure of a minimal nitroreductase ydja from escherichia2 coli k12 with and without fmn cofactor
|
| 39 | c1fdvA_ |
|
not modelled |
16.4 |
19 |
PDB header:dehydrogenase
Chain: A: PDB Molecule:17-beta-hydroxysteroid dehydrogenase;
PDBTitle: human 17-beta-hydroxysteroid-dehydrogenase type 1 mutant2 h221l complexed with nad+
|
| 40 | c2wzvB_ |
|
not modelled |
15.3 |
8 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nfnb protein;
PDBTitle: crystal structure of the fmn-dependent nitroreductase nfnb2 from mycobacterium smegmatis
|
| 41 | d1uj4a1 |
|
not modelled |
14.3 |
15 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 42 | c1miyB_ |
|
not modelled |
12.5 |
25 |
PDB header:translation, transferase
Chain: B: PDB Molecule:trna cca-adding enzyme;
PDBTitle: crystal structure of bacillus stearothermophilus cca-adding enzyme in2 complex with ctp
|
| 43 | d1jtva_ |
|
not modelled |
11.1 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 44 | d1fcqa_ |
|
not modelled |
10.3 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Bee venom hyaluronidase |
| 45 | c3cioA_ |
|
not modelled |
9.7 |
35 |
PDB header:signaling protein, transferase
Chain: A: PDB Molecule:tyrosine-protein kinase etk;
PDBTitle: the kinase domain of escherichia coli tyrosine kinase etk
|
| 46 | c3bemA_ |
|
not modelled |
9.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative nad(p)h nitroreductase ydfn;
PDBTitle: crystal structure of putative nitroreductase ydfn (2632848) from2 bacillus subtilis at 1.65 a resolution
|
| 47 | c2atmA_ |
|
not modelled |
9.6 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronoglucosaminidase;
PDBTitle: crystal structure of the recombinant allergen ves v 2
|
| 48 | c1skoA_ |
|
not modelled |
9.5 |
17 |
PDB header:signaling protein
Chain: A: PDB Molecule:mitogen-activated protein kinase kinase 1
PDBTitle: mp1-p14 complex
|
| 49 | c1fcuA_ |
|
not modelled |
9.4 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronoglucosaminidase;
PDBTitle: crystal structure (trigonal) of bee venom hyaluronidase
|
| 50 | c3emkA_ |
|
not modelled |
9.3 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose/ribitol dehydrogenase;
PDBTitle: 2.5a crystal structure of glucose/ribitol dehydrogenase2 from brucella melitensis
|
| 51 | d1h6za2 |
|
not modelled |
9.3 |
15 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
| 52 | c3mjjD_ |
|
not modelled |
9.3 |
36 |
PDB header:hydrolase
Chain: D: PDB Molecule:predicted acetamidase/formamidase;
PDBTitle: crystal structure analysis of a recombinant predicted2 acetamidase/formamidase from the thermophile thermoanaerobacter3 tengcongensis
|
| 53 | d1tvca1 |
|
not modelled |
9.1 |
20 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
| 54 | d1ka9f_ |
|
not modelled |
9.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 55 | c1vdzA_ |
|
not modelled |
8.4 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:a-type atpase subunit a;
PDBTitle: crystal structure of a-type atpase catalytic subunit a from2 pyrococcus horikoshii ot3
|
| 56 | c2pe4A_ |
|
not modelled |
8.4 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronidase-1;
PDBTitle: structure of human hyaluronidase 1, a hyaluronan hydrolyzing enzyme2 involved in tumor growth and angiogenesis
|
| 57 | d2f4la1 |
|
not modelled |
8.4 |
21 |
Fold:CUB-like
Superfamily:Acetamidase/Formamidase-like
Family:Acetamidase/Formamidase-like |
| 58 | d1i1qb_ |
|
not modelled |
8.4 |
19 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 59 | c2xrfA_ |
|
not modelled |
8.3 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:uridine phosphorylase 2;
PDBTitle: crystal structure of human uridine phosphorylase 2
|
| 60 | c3eo7A_ |
|
not modelled |
8.3 |
24 |
PDB header:flavoprotein
Chain: A: PDB Molecule:putative nitroreductase;
PDBTitle: crystal structure of a putative nitroreductase (ava_2154) from2 anabaena variabilis atcc 29413 at 1.80 a resolution
|
| 61 | c3pbiA_ |
|
not modelled |
8.2 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:invasion protein;
PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
|
| 62 | d1h5ya_ |
|
not modelled |
8.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 63 | c3hj9A_ |
|
not modelled |
8.2 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of a putative nitroreductase (reut_a1228) from2 ralstonia eutropha jmp134 at 2.00 a resolution
|
| 64 | c3b9tD_ |
|
not modelled |
7.8 |
29 |
PDB header:hydrolase
Chain: D: PDB Molecule:twin-arginine translocation pathway signal protein;
PDBTitle: crystal structure of predicted acetamidase/formamidase (yp_546212.1)2 from methylobacillus flagellatus kt at 1.58 a resolution
|
| 65 | c2xivA_ |
|
not modelled |
7.7 |
26 |
PDB header:structural protein
Chain: A: PDB Molecule:hypothetical invasion protein;
PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
|
| 66 | c2r01A_ |
|
not modelled |
7.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nitroreductase family protein;
PDBTitle: crystal structure of a putative fmn-dependent nitroreductase (ct0345)2 from chlorobium tepidum tls at 1.15 a resolution
|
| 67 | d2fsua1 |
|
not modelled |
7.0 |
10 |
Fold:PLP-dependent transferase-like
Superfamily:PhnH-like
Family:PhnH-like |
| 68 | c2fsuA_ |
|
not modelled |
7.0 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein phnh;
PDBTitle: crystal structure of the phnh protein from escherichia coli
|
| 69 | d1eara1 |
|
not modelled |
7.0 |
30 |
Fold:Urease metallochaperone UreE, N-terminal domain
Superfamily:Urease metallochaperone UreE, N-terminal domain
Family:Urease metallochaperone UreE, N-terminal domain |
| 70 | d1j5ya1 |
|
not modelled |
6.8 |
25 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Biotin repressor-like |
| 71 | c2i7hE_ |
|
not modelled |
6.7 |
10 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:nitroreductase-like family protein;
PDBTitle: crystal structure of the nitroreductase-like family protein from2 bacillus cereus
|
| 72 | c2ii1A_ |
|
not modelled |
6.6 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:acetamidase;
PDBTitle: crystal structure of acetamidase (10172637) from bacillus halodurans2 at 1.95 a resolution
|
| 73 | c3gt2A_ |
|
not modelled |
6.6 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
|
| 74 | c3rkuC_ |
|
not modelled |
6.5 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase ymr226c;
PDBTitle: substrate fingerprint and the structure of nadp+ dependent serine2 dehydrogenase from saccharomyces cerevisiae complexed with nadp+
|
| 75 | c1vbhA_ |
|
not modelled |
6.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate,orthophosphate dikinase;
PDBTitle: pyruvate phosphate dikinase with bound mg-pep from maize
|
| 76 | d2cs7a1 |
|
not modelled |
6.3 |
21 |
Fold:IL8-like
Superfamily:PhtA domain-like
Family:PhtA domain-like |
| 77 | d1gph12 |
|
not modelled |
6.1 |
16 |
Fold:Ntn hydrolase-like
Superfamily:N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family:Class II glutamine amidotransferases |
| 78 | c1y43B_ |
|
not modelled |
5.9 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:aspergillopepsin ii heavy chain;
PDBTitle: crystal structure of aspergilloglutamic peptidase from2 aspergillus niger
|
| 79 | c2kytA_ |
|
not modelled |
5.7 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:group xvi phospholipase a2;
PDBTitle: solution struture of the h-rev107 n-terminal domain
|
| 80 | c3k81D_ |
|
not modelled |
5.7 |
25 |
PDB header:immune system, rna binding protein
Chain: D: PDB Molecule:mp18 rna editing complex protein;
PDBTitle: structure of the central interaction protein from the trypanosoma2 brucei editosome in complex with single domain antibodies
|
| 81 | d1vzwa1 |
|
not modelled |
5.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 82 | d1p3da2 |
|
not modelled |
5.5 |
27 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 83 | c3ny0D_ |
|
not modelled |
5.5 |
20 |
PDB header:metal binding protein
Chain: D: PDB Molecule:urease accessory protein uree;
PDBTitle: crystal structure of uree from helicobacter pylori (ni2+ bound form)
|
| 84 | c2wknE_ |
|
not modelled |
5.3 |
36 |
PDB header:hydrolase
Chain: E: PDB Molecule:formamidase;
PDBTitle: gamma lactamase from delftia acidovorans
|
| 85 | d1vkya_ |
|
not modelled |
5.2 |
18 |
Fold:QueA-like
Superfamily:QueA-like
Family:QueA-like |