| 1 | c3o3nB_ |
|
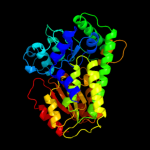 |
100.0 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:beta-subunit 2-hydroxyacyl-coa dehydratase;
PDBTitle: (r)-2-hydroxyisocaproyl-coa dehydratase in complex with its substrate2 (r)-2-hydroxyisocaproyl-coa
|
| 2 | c3o3nA_ |
|
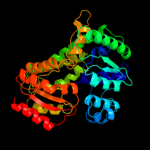 |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:alpha-subunit 2-hydroxyisocaproyl-coa dehydratase;
PDBTitle: (r)-2-hydroxyisocaproyl-coa dehydratase in complex with its substrate2 (r)-2-hydroxyisocaproyl-coa
|
| 3 | c2dwcB_ |
|
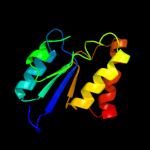 |
91.4 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:433aa long hypothetical phosphoribosylglycinamide formyl
PDBTitle: crystal structure of probable phosphoribosylglycinamide formyl2 transferase from pyrococcus horikoshii ot3 complexed with adp
|
| 4 | c3ouzA_ |
|
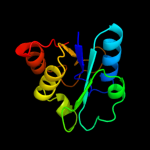 |
90.4 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of biotin carboxylase-adp complex from campylobacter2 jejuni
|
| 5 | c2vpqA_ |
|
 |
89.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coa carboxylase;
PDBTitle: crystal structure of biotin carboxylase from s. aureus2 complexed with amppnp
|
| 6 | c3g8cB_ |
|
 |
87.4 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:biotin carboxylase;
PDBTitle: crystal stucture of biotin carboxylase in complex with2 biotin, bicarbonate, adp and mg ion
|
| 7 | c2gpwC_ |
|
 |
87.4 |
18 |
PDB header:ligase
Chain: C: PDB Molecule:biotin carboxylase;
PDBTitle: crystal structure of the biotin carboxylase subunit, f363a2 mutant, of acetyl-coa carboxylase from escherichia coli.
|
| 8 | d1m1nb_ |
|
 |
86.6 |
15 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 9 | c1kjjA_ |
|
 |
86.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase 2;
PDBTitle: crystal structure of glycniamide ribonucleotide2 transformylase in complex with mg-atp-gamma-s
|
| 10 | c2pn1A_ |
|
 |
86.1 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoylphosphate synthase large subunit;
PDBTitle: crystal structure of carbamoylphosphate synthase large subunit (split2 gene in mj) (zp_00538348.1) from exiguobacterium sp. 255-15 at 2.00 a3 resolution
|
| 11 | c1ulzA_ |
|
 |
83.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase n-terminal domain;
PDBTitle: crystal structure of the biotin carboxylase subunit of pyruvate2 carboxylase
|
| 12 | c3pshA_ |
|
 |
82.8 |
14 |
PDB header:metal transport
Chain: A: PDB Molecule:protein hi_1472;
PDBTitle: classification of a haemophilus influenzae abc transporter hi1470/712 through its cognate molybdate periplasmic binding protein mola (mola3 bound to molybdate)
|
| 13 | d1n2za_ |
|
 |
81.4 |
13 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TroA-like |
| 14 | c3pdiB_ |
|
 |
79.4 |
14 |
PDB header:protein binding
Chain: B: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nifn;
PDBTitle: precursor bound nifen
|
| 15 | d1wo8a1 |
|
 |
78.0 |
20 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 16 | c2dzdB_ |
|
 |
76.1 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of the biotin carboxylase domain of2 pyruvate carboxylase
|
| 17 | d1nu0a_ |
|
 |
73.9 |
6 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Putative Holliday junction resolvase RuvX |
| 18 | c2r79A_ |
|
 |
73.7 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic binding protein;
PDBTitle: crystal structure of a periplasmic heme binding protein from2 pseudomonas aeruginosa
|
| 19 | c1m6vE_ |
|
 |
73.0 |
19 |
PDB header:ligase
Chain: E: PDB Molecule:carbamoyl phosphate synthetase large chain;
PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
|
| 20 | d1qh8b_ |
|
 |
71.0 |
15 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 21 | c3gidB_ |
|
not modelled |
68.0 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: the biotin carboxylase (bc) domain of human acetyl-coa2 carboxylase 2 (acc2) in complex with soraphen a
|
| 22 | c3u9sE_ |
|
not modelled |
67.8 |
15 |
PDB header:ligase
Chain: E: PDB Molecule:methylcrotonyl-coa carboxylase, alpha-subunit;
PDBTitle: crystal structure of p. aeruginosa 3-methylcrotonyl-coa carboxylase2 (mcc) 750 kd holoenzyme, coa complex
|
| 23 | c2vlbC_ |
|
not modelled |
67.6 |
11 |
PDB header:lyase
Chain: C: PDB Molecule:arylmalonate decarboxylase;
PDBTitle: structure of unliganded arylmalonate decarboxylase
|
| 24 | d1m1na_ |
|
not modelled |
67.0 |
13 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 25 | c2dgdD_ |
|
not modelled |
66.1 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:223aa long hypothetical arylmalonate decarboxylase;
PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
|
| 26 | d1qh8a_ |
|
not modelled |
63.5 |
13 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 27 | d1pq4a_ |
|
not modelled |
63.2 |
15 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TroA-like |
| 28 | c2xdqA_ |
|
not modelled |
61.2 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:light-independent protochlorophyllide reductase subunit n;
PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-2 chlb)2 complex
|
| 29 | d1a9xa4 |
|
not modelled |
59.9 |
19 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 30 | d1tr9a_ |
|
not modelled |
57.3 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:NagZ-like |
| 31 | c3qg5D_ |
|
not modelled |
57.3 |
10 |
PDB header:hydrolase
Chain: D: PDB Molecule:mre11;
PDBTitle: the mre11:rad50 complex forms an atp dependent molecular clamp in dna2 double-strand break repair
|
| 32 | c2xecD_ |
|
not modelled |
55.9 |
20 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
| 33 | c2yvqA_ |
|
not modelled |
55.2 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
| 34 | d2phza1 |
|
not modelled |
54.6 |
11 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:TM0189-like |
| 35 | c2phzA_ |
|
not modelled |
54.6 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:iron-uptake system-binding protein;
PDBTitle: crystal structure of iron-uptake system-binding protein2 feua from bacillus subtilis. northeast structural genomics3 target sr580.
|
| 36 | d1uf3a_ |
|
not modelled |
54.6 |
8 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TT1561-like |
| 37 | d1vhxa_ |
|
not modelled |
54.3 |
20 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Putative Holliday junction resolvase RuvX |
| 38 | c3bg5C_ |
|
not modelled |
54.1 |
16 |
PDB header:ligase
Chain: C: PDB Molecule:pyruvate carboxylase;
PDBTitle: crystal structure of staphylococcus aureus pyruvate2 carboxylase
|
| 39 | c3opyH_ |
|
not modelled |
53.8 |
20 |
PDB header:transferase
Chain: H: PDB Molecule:6-phosphofructo-1-kinase beta-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
| 40 | c3opyB_ |
|
not modelled |
53.8 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:6-phosphofructo-1-kinase beta-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
| 41 | c3s81A_ |
|
not modelled |
52.6 |
9 |
PDB header:isomerase
Chain: A: PDB Molecule:putative aspartate racemase;
PDBTitle: crystal structure of putative aspartate racemase from salmonella2 typhimurium
|
| 42 | c3ke8A_ |
|
not modelled |
51.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate
PDBTitle: crystal structure of isph:hmbpp-complex
|
| 43 | c2r7aC_ |
|
not modelled |
51.0 |
14 |
PDB header:transport protein
Chain: C: PDB Molecule:bacterial heme binding protein;
PDBTitle: crystal structure of a periplasmic heme binding protein2 from shigella dysenteriae
|
| 44 | c2ip4A_ |
|
not modelled |
50.0 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: crystal structure of glycinamide ribonucleotide synthetase from2 thermus thermophilus hb8
|
| 45 | d2ajta2 |
|
not modelled |
48.8 |
10 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:AraA N-terminal and middle domain-like |
| 46 | d1iv0a_ |
|
not modelled |
48.2 |
9 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Putative Holliday junction resolvase RuvX |
| 47 | c2hjwA_ |
|
not modelled |
47.6 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:acetyl-coa carboxylase 2;
PDBTitle: crystal structure of the bc domain of acc2
|
| 48 | d1m0wa1 |
|
not modelled |
46.1 |
11 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:Eukaryotic glutathione synthetase, substrate-binding domain |
| 49 | d1j6xa_ |
|
not modelled |
44.7 |
27 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 50 | c3t1iC_ |
|
not modelled |
43.2 |
12 |
PDB header:hydrolase
Chain: C: PDB Molecule:double-strand break repair protein mre11a;
PDBTitle: crystal structure of human mre11: understanding tumorigenic mutations
|
| 51 | c3gfvA_ |
|
not modelled |
42.9 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized abc transporter solute-binding
PDBTitle: crystal structure of petrobactin-binding protein yclq from2 bacillu subtilis
|
| 52 | c3n6rK_ |
|
not modelled |
41.4 |
11 |
PDB header:ligase
Chain: K: PDB Molecule:propionyl-coa carboxylase, alpha subunit;
PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
|
| 53 | c3cx3A_ |
|
not modelled |
41.2 |
15 |
PDB header:metal binding protein
Chain: A: PDB Molecule:lipoprotein;
PDBTitle: crystal structure analysis of the streptococcus pneumoniae2 adcaii protein
|
| 54 | c3rxyA_ |
|
not modelled |
40.8 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nif3 protein;
PDBTitle: crystal structure of nif3 superfamily protein from sphaerobacter2 thermophilus
|
| 55 | c3lp8A_ |
|
not modelled |
38.8 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine-glycine ligase;
PDBTitle: crystal structure of phosphoribosylamine-glycine ligase from2 ehrlichia chaffeensis
|
| 56 | d2j9ga2 |
|
not modelled |
38.7 |
17 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 57 | c3md9A_ |
|
not modelled |
38.4 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:hemin-binding periplasmic protein hmut;
PDBTitle: structure of apo form of a periplasmic heme binding protein
|
| 58 | c1gsoA_ |
|
not modelled |
38.0 |
10 |
PDB header:ligase
Chain: A: PDB Molecule:protein (glycinamide ribonucleotide synthetase);
PDBTitle: glycinamide ribonucleotide synthetase (gar-syn) from e.2 coli.
|
| 59 | c2xd4A_ |
|
not modelled |
37.8 |
10 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylamine--glycine ligase;
PDBTitle: nucleotide-bound structures of bacillus subtilis glycinamide2 ribonucleotide synthetase
|
| 60 | c3dloC_ |
|
not modelled |
37.5 |
20 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:universal stress protein;
PDBTitle: structure of universal stress protein from archaeoglobus fulgidus
|
| 61 | d1a9xa2 |
|
not modelled |
37.0 |
12 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain |
| 62 | d1tq8a_ |
|
not modelled |
36.9 |
10 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 63 | d2z3va1 |
|
not modelled |
36.4 |
10 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 64 | c1fuiB_ |
|
not modelled |
36.0 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:l-fucose isomerase;
PDBTitle: l-fucose isomerase from escherichia coli
|
| 65 | d1mioa_ |
|
not modelled |
35.1 |
8 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 66 | c3eiwA_ |
|
not modelled |
35.1 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:htsa protein;
PDBTitle: crystal structure of staphylococcus aureus lipoprotein, htsa
|
| 67 | c2a7vA_ |
|
not modelled |
34.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:serine hydroxymethyltransferase;
PDBTitle: human mitochondrial serine hydroxymethyltransferase 2
|
| 68 | d2a7va1 |
|
not modelled |
34.2 |
18 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 69 | c3cqyA_ |
|
not modelled |
33.9 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:anhydro-n-acetylmuramic acid kinase;
PDBTitle: crystal structure of a functionally unknown protein (so_1313) from2 shewanella oneidensis mr-1
|
| 70 | c3qvjB_ |
|
not modelled |
33.6 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:putative hydantoin racemase;
PDBTitle: allantoin racemase from klebsiella pneumoniae
|
| 71 | d2auna1 |
|
not modelled |
33.5 |
12 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LD-carboxypeptidase A C-terminal domain-like
Family:LD-carboxypeptidase A C-terminal domain-like |
| 72 | c3kalB_ |
|
not modelled |
32.8 |
22 |
PDB header:ligase
Chain: B: PDB Molecule:homoglutathione synthetase;
PDBTitle: structure of homoglutathione synthetase from glycine max in2 closed conformation with homoglutathione, adp, a sulfate3 ion, and three magnesium ions bound
|
| 73 | c2ys6A_ |
|
not modelled |
32.8 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:phosphoribosylglycinamide synthetase;
PDBTitle: crystal structure of gar synthetase from geobacillus kaustophilus
|
| 74 | c1w96B_ |
|
not modelled |
32.6 |
21 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase;
PDBTitle: crystal structure of biotin carboxylase domain of acetyl-2 coenzyme a carboxylase from saccharomyces cerevisiae in3 complex with soraphen a
|
| 75 | c3aerB_ |
|
not modelled |
32.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
|
| 76 | c2ixaA_ |
|
not modelled |
31.9 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylgalactosaminidase;
PDBTitle: a-zyme, n-acetylgalactosaminidase
|
| 77 | c2ov3A_ |
|
not modelled |
31.3 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic binding protein component of an abc
PDBTitle: crystal structure of 138-173 znua deletion mutant plus zinc2 bound
|
| 78 | c3opyE_ |
|
not modelled |
31.2 |
16 |
PDB header:transferase
Chain: E: PDB Molecule:6-phosphofructo-1-kinase alpha-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
| 79 | c1m0tB_ |
|
not modelled |
31.2 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:glutathione synthetase;
PDBTitle: yeast glutathione synthase
|
| 80 | c3hjtB_ |
|
not modelled |
31.1 |
15 |
PDB header:cell adhesion, transport protein
Chain: B: PDB Molecule:lmb;
PDBTitle: structure of laminin binding protein (lmb) of streptococcus2 agalactiae a bifunctional protein with adhesin and metal3 transporting activity
|
| 81 | c3hgmD_ |
|
not modelled |
30.8 |
10 |
PDB header:signaling protein
Chain: D: PDB Molecule:universal stress protein tead;
PDBTitle: universal stress protein tead from the trap transporter2 teaabc of halomonas elongata
|
| 82 | c2qk4A_ |
|
not modelled |
30.6 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:trifunctional purine biosynthetic protein adenosine-3;
PDBTitle: human glycinamide ribonucleotide synthetase
|
| 83 | c3g23A_ |
|
not modelled |
30.1 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:ld-carboxypeptidase a;
PDBTitle: crystal structure of a ld-carboxypeptidase a (saro_1426) from2 novosphingobium aromaticivorans dsm at 1.89 a resolution
|
| 84 | c2xdqB_ |
|
not modelled |
29.7 |
6 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-2 chlb)2 complex
|
| 85 | d1w3ia_ |
|
not modelled |
29.1 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 86 | c3m4xA_ |
|
not modelled |
29.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:nol1/nop2/sun family protein;
PDBTitle: structure of a ribosomal methyltransferase
|
| 87 | d1kjqa2 |
|
not modelled |
28.4 |
16 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 88 | c2q8pA_ |
|
not modelled |
28.1 |
10 |
PDB header:metal transport
Chain: A: PDB Molecule:iron-regulated surface determinant e;
PDBTitle: crystal structure of selenomethionine labelled s. aureus isde2 complexed with heme
|
| 89 | c3gv0A_ |
|
not modelled |
27.7 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of laci family transcription regulator from2 agrobacterium tumefaciens
|
| 90 | c2xmoB_ |
|
not modelled |
27.7 |
7 |
PDB header:hydrolase
Chain: B: PDB Molecule:lmo2642 protein;
PDBTitle: the crystal structure of lmo2642
|
| 91 | d1vmda_ |
|
not modelled |
27.2 |
20 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 92 | c3q9cF_ |
|
not modelled |
26.9 |
11 |
PDB header:hydrolase
Chain: F: PDB Molecule:acetylpolyamine amidohydrolase;
PDBTitle: crystal structure of h159a apah complexed with n8-acetylspermidine
|
| 93 | c3ovkD_ |
|
not modelled |
26.9 |
17 |
PDB header:hydrolase
Chain: D: PDB Molecule:aminopeptidase p, xaa-pro dipeptidase;
PDBTitle: crystal structure of an xxa-pro aminopeptidase from streptococcus2 pyogenes
|
| 94 | c3opyG_ |
|
not modelled |
26.8 |
16 |
PDB header:transferase
Chain: G: PDB Molecule:6-phosphofructo-1-kinase alpha-subunit;
PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
|
| 95 | c3m6wA_ |
|
not modelled |
26.5 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:rrna methylase;
PDBTitle: multi-site-specific 16s rrna methyltransferase rsmf from thermus2 thermophilus in space group p21212 in complex with s-adenosyl-l-3 methionine
|
| 96 | c3qocD_ |
|
not modelled |
26.3 |
24 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative metallopeptidase;
PDBTitle: crystal structure of n-terminal domain (creatinase/prolidase like2 domain) of putative metallopeptidase from corynebacterium diphtheriae
|
| 97 | c1b74A_ |
|
not modelled |
26.3 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: glutamate racemase from aquifex pyrophilus
|
| 98 | c3r1fO_ |
|
not modelled |
26.1 |
16 |
PDB header:transcription
Chain: O: PDB Molecule:esx-1 secretion-associated regulator espr;
PDBTitle: crystal structure of a key regulator of virulence in mycobacterium2 tuberculosis
|
| 99 | c3il0B_ |
|
not modelled |
25.7 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:aminopeptidase p; xaa-pro aminopeptidase;
PDBTitle: the crystal structure of the aminopeptidase p,xaa-pro aminopeptidase2 from streptococcus thermophilus
|
| 100 | d1lfpa_ |
|
not modelled |
25.6 |
11 |
Fold:YebC-like
Superfamily:YebC-like
Family:YebC-like |
| 101 | c1k97A_ |
|
not modelled |
25.5 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:argininosuccinate synthase;
PDBTitle: crystal structure of e. coli argininosuccinate synthetase in complex2 with aspartate and citrulline
|
| 102 | d1pv8a_ |
|
not modelled |
25.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
| 103 | c2nytB_ |
|
not modelled |
25.4 |
8 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable c->u-editing enzyme apobec-2;
PDBTitle: the apobec2 crystal structure and functional implications2 for aid
|
| 104 | d1jvna1 |
|
not modelled |
25.2 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 105 | c1zrsB_ |
|
not modelled |
25.1 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: wild-type ld-carboxypeptidase
|
| 106 | c3pn9C_ |
|
not modelled |
25.0 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:proline dipeptidase;
PDBTitle: crystal structure of a proline dipeptidase from streptococcus2 pneumoniae tigr4
|
| 107 | c2jfqA_ |
|
not modelled |
24.6 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of staphylococcus aureus glutamate2 racemase in complex with d-glutamate
|
| 108 | c1lw7A_ |
|
not modelled |
24.4 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:transcriptional regulator nadr;
PDBTitle: nadr protein from haemophilus influenzae
|
| 109 | c2k4mA_ |
|
not modelled |
23.7 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0146 protein mth_1000;
PDBTitle: solution nmr structure of m. thermoautotrophicum protein2 mth_1000, northeast structural genomics consortium target3 tr8
|
| 110 | d3bofa2 |
|
not modelled |
23.5 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 111 | c3menC_ |
|
not modelled |
23.3 |
10 |
PDB header:hydrolase
Chain: C: PDB Molecule:acetylpolyamine aminohydrolase;
PDBTitle: crystal structure of acetylpolyamine aminohydrolase from burkholderia2 pseudomallei, iodide soak
|
| 112 | c3dx5A_ |
|
not modelled |
23.2 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
| 113 | c3av0A_ |
|
not modelled |
22.7 |
11 |
PDB header:recombination
Chain: A: PDB Molecule:dna double-strand break repair protein mre11;
PDBTitle: crystal structure of mre11-rad50 bound to atp s
|
| 114 | d1chma1 |
|
not modelled |
22.5 |
12 |
Fold:Ribonuclease H-like motif
Superfamily:Creatinase/prolidase N-terminal domain
Family:Creatinase/prolidase N-terminal domain |
| 115 | c1zp9A_ |
|
not modelled |
22.4 |
4 |
PDB header:transferase
Chain: A: PDB Molecule:rio1 kinase;
PDBTitle: crystal structure of full-legnth a.fulgidus rio1 serine kinase bound2 to atp and mn2+ ions.
|
| 116 | d1t6ca1 |
|
not modelled |
22.2 |
20 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Ppx/GppA phosphatase |
| 117 | c2frxD_ |
|
not modelled |
22.0 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:hypothetical protein yebu;
PDBTitle: crystal structure of yebu, a m5c rna methyltransferase from e.coli
|
| 118 | c2jvrA_ |
|
not modelled |
22.0 |
17 |
PDB header:rna binding protein
Chain: A: PDB Molecule:nucleolar protein 3;
PDBTitle: segmental isotope labeling of npl3p
|
| 119 | d2d13a1 |
|
not modelled |
21.9 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:N-type ATP pyrophosphatases |
| 120 | c3hfrA_ |
|
not modelled |
21.9 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
|

