| 1 | c2rjgC_ |
|
 |
100.0 |
97 |
PDB header:isomerase
Chain: C: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of biosynthetic alaine racemase from escherichia2 coli
|
| 2 | c1niuA_ |
|
 |
100.0 |
31 |
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase;
PDBTitle: alanine racemase with bound inhibitor derived from l-2 cycloserine
|
| 3 | c3mubB_ |
|
 |
100.0 |
31 |
PDB header:isomerase
Chain: B: PDB Molecule:alanine racemase;
PDBTitle: the crystal structure of alanine racemase from streptococcus2 pneumoniae
|
| 4 | c2odoC_ |
|
 |
100.0 |
46 |
PDB header:isomerase
Chain: C: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of pseudomonas fluorescens alanine racemase
|
| 5 | c3kw3B_ |
|
 |
100.0 |
29 |
PDB header:isomerase
Chain: B: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of alanine racemase from bartonella henselae with2 covalently bound pyridoxal phosphate
|
| 6 | c3e6eC_ |
|
 |
100.0 |
33 |
PDB header:isomerase
Chain: C: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of alanine racemase from e.faecalis2 complex with cycloserine
|
| 7 | c3oo2A_ |
|
 |
100.0 |
34 |
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase 1;
PDBTitle: 2.37 angstrom resolution crystal structure of an alanine racemase2 (alr) from staphylococcus aureus subsp. aureus col
|
| 8 | c1vftA_ |
|
 |
100.0 |
36 |
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of l-cycloserine-bound form of alanine2 racemase from d-cycloserine-producing streptomyces3 lavendulae
|
| 9 | c3hurA_ |
|
 |
100.0 |
29 |
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of alanine racemase from oenococcus oeni
|
| 10 | c3co8B_ |
|
 |
100.0 |
27 |
PDB header:isomerase
Chain: B: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of alanine racemase from oenococcus oeni
|
| 11 | c3oo2B_ |
|
 |
100.0 |
33 |
PDB header:isomerase
Chain: B: PDB Molecule:alanine racemase 1;
PDBTitle: 2.37 angstrom resolution crystal structure of an alanine racemase2 (alr) from staphylococcus aureus subsp. aureus col
|
| 12 | c2vd9A_ |
|
 |
100.0 |
30 |
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase;
PDBTitle: the crystal structure of alanine racemase from bacillus2 anthracis (ba0252) with bound l-ala-p
|
| 13 | c2dy3B_ |
|
 |
100.0 |
33 |
PDB header:isomerase
Chain: B: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of alanine racemase from corynebacterium glutamicum
|
| 14 | c1xfcB_ |
|
 |
100.0 |
36 |
PDB header:isomerase
Chain: B: PDB Molecule:alanine racemase;
PDBTitle: the 1.9 a crystal structure of alanine racemase from mycobacterium2 tuberculosis contains a conserved entryway into the active site
|
| 15 | d1rcqa2 |
|
 |
100.0 |
45 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
| 16 | d1bd0a2 |
|
 |
100.0 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
| 17 | d1vfsa2 |
|
 |
100.0 |
35 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
| 18 | d1rcqa1 |
|
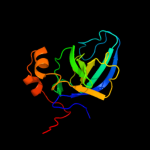 |
100.0 |
45 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Alanine racemase |
| 19 | d1vfsa1 |
|
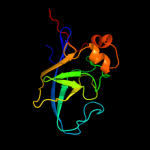 |
100.0 |
38 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Alanine racemase |
| 20 | d1bd0a1 |
|
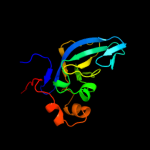 |
100.0 |
31 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Alanine racemase |
| 21 | c3anuA_ |
|
not modelled |
100.0 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:d-serine dehydratase;
PDBTitle: crystal structure of d-serine dehydratase from chicken kidney
|
| 22 | c3gwqB_ |
|
not modelled |
100.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:d-serine deaminase;
PDBTitle: crystal structure of a putative d-serine deaminase (bxe_a4060) from2 burkholderia xenovorans lb400 at 2.00 a resolution
|
| 23 | c1njjC_ |
|
not modelled |
100.0 |
11 |
PDB header:lyase
Chain: C: PDB Molecule:ornithine decarboxylase;
PDBTitle: crystal structure determination of t. brucei ornithine2 decarboxylase bound to d-ornithine and to g418
|
| 24 | c2j66A_ |
|
not modelled |
100.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:btrk;
PDBTitle: structural characterisation of btrk decarboxylase from2 butirosin biosynthesis
|
| 25 | c2p3eA_ |
|
not modelled |
100.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of aq1208 from aquifex aeolicus
|
| 26 | c3llxA_ |
|
not modelled |
100.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:predicted amino acid aldolase or racemase;
PDBTitle: crystal structure of an ala racemase-like protein (il1761) from2 idiomarina loihiensis at 1.50 a resolution
|
| 27 | c2qghA_ |
|
not modelled |
100.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase from helicobacter2 pylori complexed with l-lysine
|
| 28 | c3r79B_ |
|
not modelled |
100.0 |
16 |
PDB header:structure genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an uncharactertized protein from agrobacterium2 tumefaciens
|
| 29 | c1w8gA_ |
|
not modelled |
100.0 |
12 |
PDB header:plp-binding protein
Chain: A: PDB Molecule:hypothetical upf0001 protein yggs;
PDBTitle: crystal structure of e. coli k-12 yggs
|
| 30 | c3n2bD_ |
|
not modelled |
100.0 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: 1.8 angstrom resolution crystal structure of diaminopimelate2 decarboxylase (lysa) from vibrio cholerae.
|
| 31 | c1tufA_ |
|
not modelled |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase from m.2 jannaschi
|
| 32 | d1ct5a_ |
|
not modelled |
100.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:"Hypothetical" protein ybl036c |
| 33 | c2nvaH_ |
|
not modelled |
100.0 |
14 |
PDB header:lyase
Chain: H: PDB Molecule:arginine decarboxylase, a207r protein;
PDBTitle: the x-ray crystal structure of the paramecium bursaria2 chlorella virus arginine decarboxylase bound to agmatine
|
| 34 | c3cpgA_ |
|
not modelled |
100.0 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an unknown protein from bifidobacterium2 adolescentis
|
| 35 | c2pljA_ |
|
not modelled |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:lysine/ornithine decarboxylase;
PDBTitle: crystal structure of lysine/ornithine decarboxylase complexed with2 putrescine from vibrio vulnificus
|
| 36 | c2yxxA_ |
|
not modelled |
100.0 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure analysis of diaminopimelate decarboxylate (lysa)
|
| 37 | c2o0tB_ |
|
not modelled |
100.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: the three dimensional structure of diaminopimelate decarboxylase from2 mycobacterium tuberculosis reveals a tetrameric enzyme organisation
|
| 38 | c1knwA_ |
|
not modelled |
100.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:diaminopimelate decarboxylase;
PDBTitle: crystal structure of diaminopimelate decarboxylase
|
| 39 | c2on3A_ |
|
not modelled |
99.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:ornithine decarboxylase;
PDBTitle: a structural insight into the inhibition of human and2 leishmania donovani ornithine decarboxylases by 3-aminooxy-3 1-aminopropane
|
| 40 | c3btnA_ |
|
not modelled |
99.9 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:antizyme inhibitor 1;
PDBTitle: crystal structure of antizyme inhibitor, an ornithine2 decarboxylase homologous protein
|
| 41 | d1f3ta2 |
|
not modelled |
99.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
| 42 | c3n29A_ |
|
not modelled |
99.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:carboxynorspermidine decarboxylase;
PDBTitle: crystal structure of carboxynorspermidine decarboxylase complexed with2 norspermidine from campylobacter jejuni
|
| 43 | c1d7kB_ |
|
not modelled |
99.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:human ornithine decarboxylase;
PDBTitle: crystal structure of human ornithine decarboxylase at 2.12 angstroms resolution
|
| 44 | d7odca2 |
|
not modelled |
99.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
| 45 | d1hkva2 |
|
not modelled |
99.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
| 46 | c3mt1B_ |
|
not modelled |
99.8 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:putative carboxynorspermidine decarboxylase protein;
PDBTitle: crystal structure of putative carboxynorspermidine decarboxylase2 protein from sinorhizobium meliloti
|
| 47 | d1d7ka2 |
|
not modelled |
99.8 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
| 48 | d1twia2 |
|
not modelled |
99.8 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
| 49 | d1knwa2 |
|
not modelled |
99.6 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
| 50 | c3nzpA_ |
|
not modelled |
99.6 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:arginine decarboxylase;
PDBTitle: crystal structure of the biosynthetic arginine decarboxylase spea from2 campylobacter jejuni, northeast structural genomics consortium target3 br53
|
| 51 | c3nzqB_ |
|
not modelled |
99.6 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:biosynthetic arginine decarboxylase;
PDBTitle: crystal structure of biosynthetic arginine decarboxylase adc (spea)2 from escherichia coli, northeast structural genomics consortium3 target er600
|
| 52 | c3n2oA_ |
|
not modelled |
99.5 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:biosynthetic arginine decarboxylase;
PDBTitle: x-ray crystal structure of arginine decarboxylase complexed with2 arginine from vibrio vulnificus
|
| 53 | d1y0ya1 |
|
not modelled |
69.7 |
27 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Aminopeptidase/glucanase lid domain
Family:Aminopeptidase/glucanase lid domain |
| 54 | d2grea1 |
|
not modelled |
66.5 |
22 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Aminopeptidase/glucanase lid domain
Family:Aminopeptidase/glucanase lid domain |
| 55 | d1knwa1 |
|
not modelled |
58.7 |
25 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 56 | d1twia1 |
|
not modelled |
54.9 |
17 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 57 | c3c2qA_ |
|
not modelled |
49.1 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized conserved protein;
PDBTitle: crystal structure of conserved putative lor/sdh protein2 from methanococcus maripaludis s2
|
| 58 | d2toda1 |
|
not modelled |
32.4 |
13 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 59 | d1yloa1 |
|
not modelled |
25.1 |
27 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Aminopeptidase/glucanase lid domain
Family:Aminopeptidase/glucanase lid domain |
| 60 | d1njjb1 |
|
not modelled |
22.2 |
13 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 61 | d1hkva1 |
|
not modelled |
17.7 |
24 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 62 | d1vhea1 |
|
not modelled |
17.3 |
17 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Aminopeptidase/glucanase lid domain
Family:Aminopeptidase/glucanase lid domain |
| 63 | d1f3ta1 |
|
not modelled |
17.0 |
16 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 64 | c2l55A_ |
|
not modelled |
17.0 |
31 |
PDB header:metal binding protein
Chain: A: PDB Molecule:silb,silver efflux protein, mfp component of the three
PDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
|
| 65 | c2l8kA_ |
|
not modelled |
16.9 |
31 |
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 7;
PDBTitle: nmr structure of the arterivirus nonstructural protein 7 alpha (nsp72 alpha)
|
| 66 | d1vqoe2 |
|
not modelled |
16.4 |
19 |
Fold:Ribosomal protein L6
Superfamily:Ribosomal protein L6
Family:Ribosomal protein L6 |
| 67 | c1uheA_ |
|
not modelled |
16.2 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:aspartate 1-decarboxylase alpha chain;
PDBTitle: crystal structure of aspartate decarboxylase, isoaspargine complex
|
| 68 | d2fvga1 |
|
not modelled |
15.9 |
24 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Aminopeptidase/glucanase lid domain
Family:Aminopeptidase/glucanase lid domain |
| 69 | c1zeqX_ |
|
not modelled |
15.7 |
19 |
PDB header:metal binding protein
Chain: X: PDB Molecule:cation efflux system protein cusf;
PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
|
| 70 | d2cbia2 |
|
not modelled |
15.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:alpha-D-glucuronidase/Hyaluronidase catalytic domain |
| 71 | d1vcta2 |
|
not modelled |
12.7 |
17 |
Fold:TrkA C-terminal domain-like
Superfamily:TrkA C-terminal domain-like
Family:TrkA C-terminal domain-like |
| 72 | d1x6oa1 |
|
not modelled |
12.6 |
32 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 73 | d2g46a1 |
|
not modelled |
12.3 |
24 |
Fold:beta-clip
Superfamily:SET domain
Family:Viral histone H3 Lysine 27 Methyltransferase |
| 74 | d2q07a1 |
|
not modelled |
10.3 |
10 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:PUA domain |
| 75 | c3b8iF_ |
|
not modelled |
10.1 |
15 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
| 76 | d1vhoa1 |
|
not modelled |
9.4 |
29 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Aminopeptidase/glucanase lid domain
Family:Aminopeptidase/glucanase lid domain |
| 77 | d1h3ia2 |
|
not modelled |
9.3 |
29 |
Fold:beta-clip
Superfamily:SET domain
Family:Histone lysine methyltransferases |
| 78 | c3rnvA_ |
|
not modelled |
8.9 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:helper component proteinase;
PDBTitle: structure of the autocatalytic cysteine protease domain of potyvirus2 helper-component proteinase
|
| 79 | d1g94a2 |
|
not modelled |
8.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 80 | d1i7aa_ |
|
not modelled |
8.8 |
7 |
Fold:PH domain-like barrel
Superfamily:PH domain-like
Family:Enabled/VASP homology 1 domain (EVH1 domain) |
| 81 | d1bkba1 |
|
not modelled |
8.6 |
21 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 82 | c2bknA_ |
|
not modelled |
8.5 |
16 |
PDB header:membrane protein
Chain: A: PDB Molecule:hypothetical protein ph0236;
PDBTitle: structure analysis of unknown function protein
|
| 83 | c1pkvB_ |
|
not modelled |
8.3 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: the n-terminal domain of riboflavin synthase in complex with2 riboflavin
|
| 84 | c1pkvA_ |
|
not modelled |
8.3 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:riboflavin synthase alpha chain;
PDBTitle: the n-terminal domain of riboflavin synthase in complex with2 riboflavin
|
| 85 | d2eifa1 |
|
not modelled |
8.2 |
26 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 86 | d1khia1 |
|
not modelled |
8.0 |
16 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 87 | d1iz6a1 |
|
not modelled |
7.6 |
16 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 88 | c3zvrA_ |
|
not modelled |
7.5 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:dynamin-1;
PDBTitle: crystal structure of dynamin
|
| 89 | c2zp2B_ |
|
not modelled |
7.5 |
28 |
PDB header:transferase inhibitor
Chain: B: PDB Molecule:kinase a inhibitor;
PDBTitle: c-terminal domain of kipi from bacillus subtilis
|
| 90 | d1j98a_ |
|
not modelled |
6.8 |
21 |
Fold:LuxS/MPP-like metallohydrolase
Superfamily:LuxS/MPP-like metallohydrolase
Family:Autoinducer-2 production protein LuxS |
| 91 | c3kgkA_ |
|
not modelled |
6.8 |
7 |
PDB header:chaperone
Chain: A: PDB Molecule:arsenical resistance operon trans-acting repressor arsd;
PDBTitle: crystal structure of arsd
|
| 92 | d2f69a2 |
|
not modelled |
6.7 |
30 |
Fold:beta-clip
Superfamily:SET domain
Family:Histone lysine methyltransferases |
| 93 | c2vl6C_ |
|
not modelled |
6.7 |
38 |
PDB header:dna binding protein
Chain: C: PDB Molecule:minichromosome maintenance protein mcm;
PDBTitle: structural analysis of the sulfolobus solfataricus mcm2 protein n-terminal domain
|
| 94 | c3mmlD_ |
|
not modelled |
6.6 |
25 |
PDB header:hydrolase
Chain: D: PDB Molecule:allophanate hydrolase subunit 1;
PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
|
| 95 | c2fyuE_ |
|
not modelled |
6.6 |
7 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
|
| 96 | c1s1hQ_ |
|
not modelled |
6.5 |
26 |
PDB header:ribosome
Chain: Q: PDB Molecule:40s ribosomal protein s11;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
|
| 97 | d2fy8a2 |
|
not modelled |
6.5 |
24 |
Fold:TrkA C-terminal domain-like
Superfamily:TrkA C-terminal domain-like
Family:TrkA C-terminal domain-like |
| 98 | d1oioa_ |
|
not modelled |
6.4 |
21 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:Bacterial adhesins
Family:F17c-type adhesin |
| 99 | d1o75a1 |
|
not modelled |
6.4 |
43 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Tp47 lipoprotein, middle and C-terminal domains
Family:Tp47 lipoprotein, middle and C-terminal domains |

