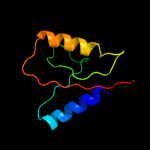
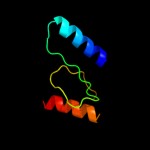
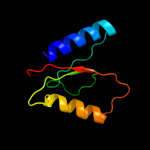
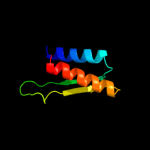
1 c3upsA_
100.0
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: iojap-like protein;PDBTitle: crystal structure of iojap-like protein from zymomonas mobilis
2 d2id1a1
100.0
36
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Iojap/YbeB-like3 d2o5aa1
100.0
36
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Iojap/YbeB-like4 c2omdB_
86.4
13
PDB header: lyaseChain: B: PDB Molecule: molybdopterin-converting factor subunit 2;PDBTitle: crystal structure of molybdopterin converting factor subunit 22 (aq_2181) from aquifex aeolicus vf5
5 c2qieA_
74.3
13
PDB header: transferaseChain: A: PDB Molecule: molybdopterin-converting factor subunit 2;PDBTitle: staphylococcus aureus molybdopterin synthase in complex2 with precursor z
6 d1fm0e_
69.9
12
Fold: alpha/beta-HammerheadSuperfamily: Molybdopterin synthase subunit MoaEFamily: Molybdopterin synthase subunit MoaE7 d1ex9a_
51.8
12
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Bacterial lipase8 d1vmea1
51.4
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related9 c3b6iB_
40.4
21
PDB header: flavoproteinChain: B: PDB Molecule: flavoprotein wrba;PDBTitle: wrba from escherichia coli, native structure
10 d1n0ua4
39.3
8
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: EF-G/eEF-2 domains III and V11 d2arka1
34.9
17
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like12 d2dy1a4
29.7
11
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: EF-G/eEF-2 domains III and V13 c3fniA_
29.7
11
PDB header: oxidoreductaseChain: A: PDB Molecule: putative diflavin flavoprotein a 3;PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
14 d2bm0a4
28.7
5
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: EF-G/eEF-2 domains III and V15 d1e5da1
28.0
12
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related16 c3hlyA_
27.4
14
PDB header: flavoproteinChain: A: PDB Molecule: flavodoxin-like domain;PDBTitle: crystal structure of the flavodoxin-like domain from2 synechococcus sp q5mzp6_synp6 protein. northeast structural3 genomics consortium target snr135d.
17 d1ycga1
25.5
5
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related18 d1nvja_
20.2
13
Fold: alpha/beta-HammerheadSuperfamily: Molybdopterin synthase subunit MoaEFamily: Molybdopterin synthase subunit MoaE19 d1k8kd1
20.2
28
Fold: Secretion chaperone-likeSuperfamily: Arp2/3 complex subunitsFamily: Arp2/3 complex subunits20 d1lk5a2
20.1
14
Fold: Ferredoxin-likeSuperfamily: D-ribose-5-phosphate isomerase (RpiA), lid domainFamily: D-ribose-5-phosphate isomerase (RpiA), lid domain21 d2fz5a1
not modelled
19.4
10
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related22 d1xpja_
not modelled
19.3
10
Fold: HAD-likeSuperfamily: HAD-likeFamily: Hypothetical protein VC023223 c2zkiH_
not modelled
18.7
15
PDB header: transcriptionChain: H: PDB Molecule: 199aa long hypothetical trp repressor bindingPDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
24 c2wp4A_
not modelled
16.8
15
PDB header: transferaseChain: A: PDB Molecule: molybdopterin-converting factor subunit 2 1;PDBTitle: crystal structure of rv3119 from mycobacterium tuberculosis
25 c3b8hA_
not modelled
15.0
8
PDB header: biosynthetic protein/transferaseChain: A: PDB Molecule: elongation factor 2;PDBTitle: structure of the eef2-exoa(e546a)-nad+ complex
26 d2fcra_
not modelled
14.9
17
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related27 d1mp9a1
not modelled
14.2
15
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain28 c2xexA_
not modelled
13.9
8
PDB header: translationChain: A: PDB Molecule: elongation factor g;PDBTitle: crystal structure of staphylococcus aureus elongation factor2 g
29 c2pjmA_
not modelled
13.4
22
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: structure of ribose 5-phosphate isomerase a from2 methanocaldococcus jannaschii
30 d2obba1
not modelled
13.0
5
Fold: HAD-likeSuperfamily: HAD-likeFamily: BT0820-like31 c3dnpA_
not modelled
12.6
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: stress response protein yhax;PDBTitle: crystal structure of stress response protein yhax from bacillus2 subtilis
32 c3f4aA_
not modelled
12.4
14
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein ygr203w;PDBTitle: structure of ygr203w, a yeast protein tyrosine phosphatase2 of the rhodanese family
33 d1llda1
not modelled
11.4
10
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like34 c1lk5C_
not modelled
11.4
14
PDB header: isomeraseChain: C: PDB Molecule: d-ribose-5-phosphate isomerase;PDBTitle: structure of the d-ribose-5-phosphate isomerase from2 pyrococcus horikoshii
35 d2z1ca1
not modelled
10.7
23
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like36 d2ot2a1
not modelled
10.7
19
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like37 c2c5iP_
not modelled
10.6
50
PDB header: protein transportChain: P: PDB Molecule: vacuolar protein sorting protein 51;PDBTitle: n-terminal domain of tlg1 complexed with n-terminus of2 vps51 in distorted conformation
38 d5nula_
not modelled
10.4
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related39 c2c5kP_
not modelled
10.3
50
PDB header: protein transportChain: P: PDB Molecule: vacuolar protein sorting protein 51;PDBTitle: n-terminal domain of tlg1 complexed with n-terminus of2 vps51
40 d1wzca1
not modelled
10.3
5
Fold: HAD-likeSuperfamily: HAD-likeFamily: Predicted hydrolases Cof41 c1vmeB_
not modelled
10.3
13
PDB header: electron transportChain: B: PDB Molecule: flavoprotein;PDBTitle: crystal structure of flavoprotein (tm0755) from thermotoga maritima at2 1.80 a resolution
42 d1e4ea1
not modelled
10.2
23
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: D-Alanine ligase N-terminal domain43 d1ydga_
not modelled
9.8
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like44 c3o2eA_
not modelled
9.4
11
PDB header: unknown functionChain: A: PDB Molecule: bola-like protein;PDBTitle: crystal structure of a bol-like protein from babesia bovis
45 c3hheA_
not modelled
9.3
15
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from bartonella2 henselae
46 c2kz0A_
not modelled
9.2
6
PDB header: transcriptionChain: A: PDB Molecule: bola family protein;PDBTitle: solution structure of a bola protein (ech_0303) from ehrlichia2 chaffeensis. seattle structural genomics center for infectious3 disease target ehcha.10365.a
47 d1t57a_
not modelled
8.7
14
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: MTH1675-like48 c2rdo7_
not modelled
8.6
13
PDB header: ribosomeChain: 7: PDB Molecule: elongation factor g;PDBTitle: 50s subunit with ef-g(gdpnp) and rrf bound
49 c2q9uB_
not modelled
8.4
8
PDB header: oxidoreductaseChain: B: PDB Molecule: a-type flavoprotein;PDBTitle: crystal structure of the flavodiiron protein from giardia2 intestinalis
50 c1ychD_
not modelled
8.1
11
PDB header: oxidoreductaseChain: D: PDB Molecule: nitric oxide reductase;PDBTitle: x-ray crystal structures of moorella thermoacetica fpra.2 novel diiron site structure and mechanistic insights into3 a scavenging nitric oxide reductase
51 c1zn0B_
not modelled
7.9
5
PDB header: translation/biosynthetic protein/rnaChain: B: PDB Molecule: elongation factor g;PDBTitle: coordinates of rrf and ef-g fitted into cryo-em map of the2 50s subunit bound with both ef-g (gdpnp) and rrf
52 c3tqtB_
not modelled
7.7
19
PDB header: ligaseChain: B: PDB Molecule: d-alanine--d-alanine ligase;PDBTitle: structure of the d-alanine-d-alanine ligase from coxiella burnetii
53 c3f6sI_
not modelled
7.7
10
PDB header: electron transportChain: I: PDB Molecule: flavodoxin;PDBTitle: desulfovibrio desulfuricans (atcc 29577) oxidized flavodoxin2 alternate conformers
54 d1nh8a2
not modelled
7.7
13
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: ATP phosphoribosyltransferase (ATP-PRTase, HisG), regulatory C-terminal domain55 c2zf8A_
not modelled
7.5
16
PDB header: structural proteinChain: A: PDB Molecule: component of sodium-driven polar flagellar motor;PDBTitle: crystal structure of moty
56 c1e5dA_
not modelled
7.4
13
PDB header: oxidoreductaseChain: A: PDB Molecule: rubredoxin\:oxygen oxidoreductase;PDBTitle: rubredoxin oxygen:oxidoreductase (roo) from anaerobe2 desulfovibrio gigas
57 d2fvta1
not modelled
7.4
19
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like58 c3cioA_
not modelled
7.3
13
PDB header: signaling protein, transferaseChain: A: PDB Molecule: tyrosine-protein kinase etk;PDBTitle: the kinase domain of escherichia coli tyrosine kinase etk
59 d2qwxa1
not modelled
7.3
26
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Quinone reductase60 d2fi9a1
not modelled
7.2
11
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like61 c2hnbA_
not modelled
7.2
7
PDB header: electron transportChain: A: PDB Molecule: protein mioc;PDBTitle: solution structure of a bacterial holo-flavodoxin
62 c3l7oB_
not modelled
7.2
15
PDB header: isomeraseChain: B: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
63 c3o3nB_
not modelled
7.1
19
PDB header: lyaseChain: B: PDB Molecule: beta-subunit 2-hydroxyacyl-coa dehydratase;PDBTitle: (r)-2-hydroxyisocaproyl-coa dehydratase in complex with its substrate2 (r)-2-hydroxyisocaproyl-coa
64 d2hd3a1
not modelled
7.0
25
Fold: OB-foldSuperfamily: EutN/CcmL-likeFamily: EutN/CcmL-like65 c3pgvB_
not modelled
6.9
26
PDB header: hydrolaseChain: B: PDB Molecule: haloacid dehalogenase-like hydrolase;PDBTitle: crystal structure of a haloacid dehalogenase-like hydrolase2 (kpn_04322) from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at3 2.39 a resolution
66 d1nh2a1
not modelled
6.9
10
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain67 c3bzqA_
not modelled
6.9
18
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: nitrogen regulatory protein p-ii;PDBTitle: high resolution crystal structure of nitrogen regulatory protein2 (rv2919c) of mycobacterium tuberculosis
68 d1j98a_
not modelled
6.9
17
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: Autoinducer-2 production protein LuxS69 d1kqfa1
not modelled
6.8
18
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Formate dehydrogenase/DMSO reductase, C-terminal domain70 c1olsB_
not modelled
6.8
10
PDB header: oxidoreductaseChain: B: PDB Molecule: 2-oxoisovalerate dehydrogenase beta subunit;PDBTitle: roles of his291-alpha and his146-beta' in the reductive2 acylation reaction catalyzed by human branched-chain3 alpha-ketoacid dehydrogenase
71 c3niwA_
not modelled
6.5
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: haloacid dehalogenase-like hydrolase;PDBTitle: crystal structure of a haloacid dehalogenase-like hydrolase from2 bacteroides thetaiotaomicron
72 d1umya_
not modelled
6.5
32
Fold: TIM beta/alpha-barrelSuperfamily: Homocysteine S-methyltransferaseFamily: Homocysteine S-methyltransferase73 d2a5la1
not modelled
6.5
10
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like74 d2bfdb2
not modelled
6.4
10
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain75 d2cfxa2
not modelled
6.4
9
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain76 d1qnaa1
not modelled
6.3
17
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain77 d1uj4a2
not modelled
6.3
12
Fold: Ferredoxin-likeSuperfamily: D-ribose-5-phosphate isomerase (RpiA), lid domainFamily: D-ribose-5-phosphate isomerase (RpiA), lid domain78 c2bm0A_
not modelled
6.3
5
PDB header: elongation factorChain: A: PDB Molecule: elongation factor g;PDBTitle: ribosomal elongation factor g (ef-g) fusidic acid resistant2 mutant t84a
79 d1nrwa_
not modelled
6.2
26
Fold: HAD-likeSuperfamily: HAD-likeFamily: Predicted hydrolases Cof80 d1nf2a_
not modelled
6.1
5
Fold: HAD-likeSuperfamily: HAD-likeFamily: Predicted hydrolases Cof81 d1x4ma1
not modelled
6.0
17
Fold: Eukaryotic type KH-domain (KH-domain type I)Superfamily: Eukaryotic type KH-domain (KH-domain type I)Family: Eukaryotic type KH-domain (KH-domain type I)82 d1ny8a_
not modelled
6.0
11
Fold: Alpha-lytic protease prodomain-likeSuperfamily: BolA-likeFamily: BolA-like83 c3gygA_
not modelled
5.8
15
PDB header: hydrolaseChain: A: PDB Molecule: ntd biosynthesis operon putative hydrolase ntdb;PDBTitle: crystal structure of yhjk (haloacid dehalogenase-like hydrolase2 protein) from bacillus subtilis
84 d3d3ra1
not modelled
5.8
19
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like85 c3d0oA_
not modelled
5.7
10
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase 1;PDBTitle: crystal structure of lactate dehydrogenase from2 staphylococcus aureus
86 c1uj6A_
not modelled
5.7
13
PDB header: isomeraseChain: A: PDB Molecule: ribose 5-phosphate isomerase;PDBTitle: crystal structure of thermus thermophilus ribose-5-phosphate isomerase2 complexed with arabinose-5-phosphate
87 d2guma1
not modelled
5.6
26
Fold: Viral glycoprotein ectodomain-likeSuperfamily: Viral glycoprotein ectodomain-likeFamily: Glycoprotein B-like88 d1m7ja2
not modelled
5.5
13
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: D-aminoacylase89 d1otga_
not modelled
5.5
13
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 5-carboxymethyl-2-hydroxymuconate isomerase (CHMI)90 c1zosE_
not modelled
5.5
14
PDB header: hydrolaseChain: E: PDB Molecule: 5'-methylthioadenosine / s-adenosylhomocysteinePDBTitle: structure of 5'-methylthionadenosine/s-adenosylhomocysteine2 nucleosidase from s. pneumoniae with a transition-state3 inhibitor mt-imma
91 c1byyA_
not modelled
5.4
36
PDB header: membrane proteinChain: A: PDB Molecule: protein (sodium channel alpha-subunit);PDBTitle: sodium channel iia inactivation gate
92 d1iowa1
not modelled
5.4
18
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: D-Alanine ligase N-terminal domain93 c3e5nA_
not modelled
5.3
20
PDB header: ligaseChain: A: PDB Molecule: d-alanine-d-alanine ligase a;PDBTitle: crystal strucutre of d-alanine-d-alanine ligase from2 xanthomonas oryzae pv. oryzae kacc10331
94 d2a6aa1
not modelled
5.1
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like95 c1xviA_
not modelled
5.1
15
PDB header: hydrolaseChain: A: PDB Molecule: putative mannosyl-3-phosphoglycerate phosphatase;PDBTitle: crystal structure of yedp, phosphatase-like domain protein2 from escherichia coli k12
96 d1xvia_
not modelled
5.1
15
Fold: HAD-likeSuperfamily: HAD-likeFamily: Predicted hydrolases Cof97 c3fojA_
not modelled
5.1
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of ssp1007 from staphylococcus2 saprophyticus subsp. saprophyticus. northeast structural3 genomics target syr101a.