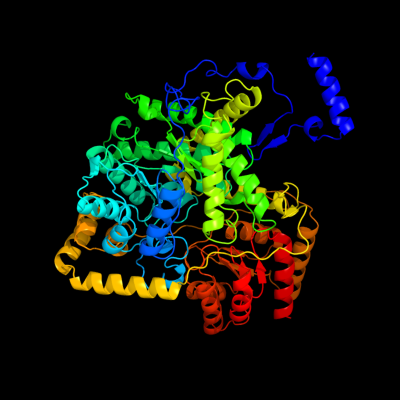
| 1 | c1e1cA_
|
|
 |
100.0 |
58 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase alpha chain;
PDBTitle: methylmalonyl-coa mutase h244a mutant
|
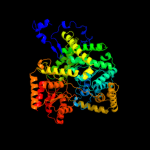
| 2 | c3bicA_
|
|
 |
100.0 |
58 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase, mitochondrial precursor;
PDBTitle: crystal structure of human methylmalonyl-coa mutase
|
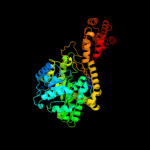
| 3 | c6reqB_
|
|
 |
100.0 |
27 |
PDB header:isomerase
Chain: B: PDB Molecule:protein (methylmalonyl-coa mutase);
PDBTitle: methylmalonyl-coa mutase, 3-carboxypropyl-coa inhibitor complex
|
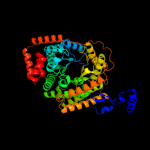
| 4 | d7reqa1
|
|
 |
100.0 |
62 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Methylmalonyl-CoA mutase, N-terminal (CoA-binding) domain |
| 5 | d7reqb1
|
|
 |
100.0 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Methylmalonyl-CoA mutase, N-terminal (CoA-binding) domain |
| 6 | d7reqb2
|
|
 |
100.0 |
16 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 7 | d7reqa2
|
|
 |
100.0 |
53 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 8 | d1ccwa_
|
|
 |
99.6 |
25 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 9 | d1xrsb1
|
|
 |
99.5 |
24 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 10 | c1xrsB_
|
|
 |
99.5 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:d-lysine 5,6-aminomutase beta subunit;
PDBTitle: crystal structure of lysine 5,6-aminomutase in complex with plp,2 cobalamin, and 5'-deoxyadenosine
|
| 11 | c2yxbA_
|
|
 |
99.4 |
42 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
| 12 | c3koxA_
|
|
 |
99.4 |
17 |
PDB header:metal binding protein
Chain: A: PDB Molecule:d-ornithine aminomutase e component;
PDBTitle: crystal structure of ornithine 4,5 aminomutase in complex with 2,4-2 diaminobutyrate (anaerobic)
|
| 13 | d1fmfa_
|
|
 |
99.1 |
24 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 14 | c1y80A_
|
|
 |
99.1 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from2 moorella thermoacetica
|
| 15 | c1bmtB_
|
|
 |
99.0 |
18 |
PDB header:methyltransferase
Chain: B: PDB Molecule:methionine synthase;
PDBTitle: how a protein binds b12: a 3.o angstrom x-ray structure of2 the b12-binding domains of methionine synthase
|
| 16 | c2i2xD_
|
|
 |
98.9 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
|
| 17 | d1ccwb_
|
|
 |
98.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Cobalamin (vitamin B12)-dependent enzymes
Family:Glutamate mutase, large subunit |
| 18 | c3ezxA_
|
|
 |
98.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:monomethylamine corrinoid protein 1;
PDBTitle: structure of methanosarcina barkeri monomethylamine2 corrinoid protein
|
| 19 | d3bula2
|
|
 |
98.7 |
17 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
| 20 | c1k98A_
|
|
 |
98.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: adomet complex of meth c-terminal fragment
|
| 21 | c1zfjA_ |
|
not modelled |
95.6 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 22 | d1krwa_ |
|
not modelled |
95.6 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 23 | c1ys7B_ |
|
not modelled |
95.6 |
18 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulatory protein prra;
PDBTitle: crystal structure of the response regulator protein prra comlexed with2 mg2+
|
| 24 | d1o4ua1 |
|
not modelled |
95.5 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 25 | c2b7pA_ |
|
not modelled |
95.5 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:probable nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of quinolinic acid phosphoribosyltransferase from2 helicobacter pylori
|
| 26 | d1yioa2 |
|
not modelled |
95.0 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 27 | d1qpoa1 |
|
not modelled |
95.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 28 | c2jbmA_ |
|
not modelled |
95.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: qprtase structure from human
|
| 29 | c1o4uA_ |
|
not modelled |
94.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:type ii quinolic acid phosphoribosyltransferase;
PDBTitle: crystal structure of a nicotinate nucleotide pyrophosphorylase2 (tm1645) from thermotoga maritima at 2.50 a resolution
|
| 30 | d1j5ta_ |
|
not modelled |
94.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 31 | d1wa3a1 |
|
not modelled |
94.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 32 | d1ys7a2 |
|
not modelled |
94.5 |
17 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 33 | c3qjaA_ |
|
not modelled |
94.1 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
|
| 34 | d1mvoa_ |
|
not modelled |
93.8 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 35 | d1zesa1 |
|
not modelled |
93.7 |
10 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 36 | c2c3zA_ |
|
not modelled |
93.6 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
| 37 | c3hdgE_ |
|
not modelled |
93.2 |
16 |
PDB header:structural genomics, unknown function
Chain: E: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the n-terminal domain of an2 uncharacterized protein (ws1339) from wolinella3 succinogenes
|
| 38 | d1u0sy_ |
|
not modelled |
93.1 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 39 | c1qpoA_ |
|
not modelled |
92.6 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:quinolinate acid phosphoribosyl transferase;
PDBTitle: quinolinate phosphoribosyl transferase (qaprtase) apo-enzyme from2 mycobacterium tuberculosis
|
| 40 | d1muma_ |
|
not modelled |
92.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 41 | d2ayxa1 |
|
not modelled |
92.5 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 42 | d1a53a_ |
|
not modelled |
92.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 43 | c3ktoA_ |
|
not modelled |
92.0 |
18 |
PDB header:transcription regulator
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of response regulator receiver protein2 from pseudoalteromonas atlantica
|
| 44 | c2yv1A_ |
|
not modelled |
91.9 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa ligase [adp-forming] subunit alpha;
PDBTitle: crystal structure of succinyl-coa synthetase alpha chain from2 methanocaldococcus jannaschii dsm 2661
|
| 45 | d1qapa1 |
|
not modelled |
91.7 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 46 | c3b8iF_ |
|
not modelled |
91.7 |
13 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
| 47 | c1x1oC_ |
|
not modelled |
91.6 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of project id tt0268 from thermus thermophilus hb8
|
| 48 | c3eooL_ |
|
not modelled |
91.6 |
16 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 49 | c2qr3A_ |
|
not modelled |
91.4 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:two-component system response regulator;
PDBTitle: crystal structure of the n-terminal signal receiver domain of two-2 component system response regulator from bacteroides fragilis
|
| 50 | c3ih1A_ |
|
not modelled |
91.3 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 51 | d1zh2a1 |
|
not modelled |
91.2 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 52 | d1dbwa_ |
|
not modelled |
91.2 |
16 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 53 | d1peya_ |
|
not modelled |
90.9 |
8 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 54 | d1i4na_ |
|
not modelled |
90.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 55 | c3jteA_ |
|
not modelled |
90.5 |
15 |
PDB header:protein binding
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of response regulator receiver domain2 protein from clostridium thermocellum
|
| 56 | d2nu7b1 |
|
not modelled |
90.4 |
15 |
Fold:Flavodoxin-like
Superfamily:Succinyl-CoA synthetase domains
Family:Succinyl-CoA synthetase domains |
| 57 | c3tdmD_ |
|
not modelled |
90.3 |
9 |
PDB header:de novo protein
Chain: D: PDB Molecule:computationally designed two-fold symmetric tim-barrel
PDBTitle: computationally designed tim-barrel protein, halfflr
|
| 58 | c2ayxA_ |
|
not modelled |
90.3 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:sensor kinase protein rcsc;
PDBTitle: solution structure of the e.coli rcsc c-terminus (residues2 700-949) containing linker region and phosphoreceiver3 domain
|
| 59 | c1gv1D_ |
|
not modelled |
90.1 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:malate dehydrogenase;
PDBTitle: structural basis for thermophilic protein stability:2 structures of thermophilic and mesophilic malate3 dehydrogenases
|
| 60 | c1ldbA_ |
|
not modelled |
89.9 |
27 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:apo-l-lactate dehydrogenase;
PDBTitle: structure determination and refinement of bacillus2 stearothermophilus lactate dehydrogenase
|
| 61 | c3eywA_ |
|
not modelled |
89.8 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
| 62 | c1ytkA_ |
|
not modelled |
89.7 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate phosphoribosyltransferase from thermoplasma
PDBTitle: crystal structure of a nicotinate phosphoribosyltransferase from2 thermoplasma acidophilum with nicotinate mononucleotide
|
| 63 | c3l0gD_ |
|
not modelled |
89.7 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: crystal structure of nicotinate-nucleotide pyrophosphorylase from2 ehrlichia chaffeensis at 2.05a resolution
|
| 64 | d1zfja1 |
|
not modelled |
89.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 65 | d2d59a1 |
|
not modelled |
89.5 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 66 | c3lyeA_ |
|
not modelled |
89.3 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 67 | c2yv2A_ |
|
not modelled |
89.3 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa synthetase alpha chain;
PDBTitle: crystal structure of succinyl-coa synthetase alpha chain from2 aeropyrum pernix k1
|
| 68 | c1zlpA_ |
|
not modelled |
89.1 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 69 | d1y81a1 |
|
not modelled |
89.0 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 70 | c3nhzA_ |
|
not modelled |
88.3 |
14 |
PDB header:dna binding protein
Chain: A: PDB Molecule:two component system transcriptional regulator mtra;
PDBTitle: structure of n-terminal domain of mtra
|
| 71 | d1y0ea_ |
|
not modelled |
88.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:NanE-like |
| 72 | d1jr1a1 |
|
not modelled |
88.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
| 73 | c2jk1A_ |
|
not modelled |
88.0 |
16 |
PDB header:dna-binding
Chain: A: PDB Molecule:hydrogenase transcriptional regulatory protein hupr1;
PDBTitle: crystal structure of the wild-type hupr receiver domain
|
| 74 | d1piia2 |
|
not modelled |
87.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 75 | d1s2wa_ |
|
not modelled |
87.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 76 | c1me9A_ |
|
not modelled |
87.8 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh) from2 tritrichomonas foetus with imp bound
|
| 77 | c3crnA_ |
|
not modelled |
87.7 |
12 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator receiver domain protein, chey-like;
PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
|
| 78 | c1ur5C_ |
|
not modelled |
87.6 |
16 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: stabilization of a tetrameric malate dehydrogenase by2 introduction of a disulfide bridge at the dimer/dimer3 interface
|
| 79 | c3pajA_ |
|
not modelled |
87.3 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase, carboxylating;
PDBTitle: 2.00 angstrom resolution crystal structure of a quinolinate2 phosphoribosyltransferase from vibrio cholerae o1 biovar eltor str.3 n16961
|
| 80 | c1e8cB_ |
|
not modelled |
87.2 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:udp-n-acetylmuramoylalanyl-d-glutamate--2,6-
PDBTitle: structure of mure the udp-n-acetylmuramyl tripeptide2 synthetase from e. coli
|
| 81 | d1ny5a1 |
|
not modelled |
87.1 |
15 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 82 | c1qapA_ |
|
not modelled |
86.9 |
19 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:quinolinic acid phosphoribosyltransferase;
PDBTitle: quinolinic acid phosphoribosyltransferase with bound2 quinolinic acid
|
| 83 | d1qkka_ |
|
not modelled |
86.7 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 84 | d1jbea_ |
|
not modelled |
86.6 |
7 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 85 | c2fpgA_ |
|
not modelled |
86.5 |
10 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa ligase [gdp-forming] alpha-chain,
PDBTitle: crystal structure of pig gtp-specific succinyl-coa2 synthetase in complex with gdp
|
| 86 | c2jrlA_ |
|
not modelled |
86.3 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator (ntrc family);
PDBTitle: solution structure of the beryllofluoride-activated ntrc4 receiver2 domain dimer
|
| 87 | c2v6bB_ |
|
not modelled |
86.2 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: crystal structure of lactate dehydrogenase from deinococcus2 radiodurans (apo form)
|
| 88 | c3fa4D_ |
|
not modelled |
86.1 |
17 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
| 89 | d1iuka_ |
|
not modelled |
86.0 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 90 | d1ujqa_ |
|
not modelled |
86.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 91 | d1thfd_ |
|
not modelled |
85.9 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 92 | d1xcfa_ |
|
not modelled |
85.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 93 | c3dl2A_ |
|
not modelled |
85.4 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ubiquitin-conjugating enzyme e2 variant 3;
PDBTitle: hexagonal structure of the ldh domain of human ubiquitin-2 conjugating enzyme e2-like isoform a
|
| 94 | c3rqiA_ |
|
not modelled |
85.3 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator protein;
PDBTitle: crystal structure of a response regulator protein from burkholderia2 pseudomallei with a phosphorylated aspartic acid, calcium ion and3 citrate
|
| 95 | c2v82A_ |
|
not modelled |
85.3 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
| 96 | d2pl1a1 |
|
not modelled |
84.9 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 97 | c8ldhA_ |
|
not modelled |
84.7 |
20 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:m4 apo-lactate dehydrogenase;
PDBTitle: refined crystal structure of dogfish m4 apo-lactate2 dehydrogenase
|
| 98 | d1rd5a_ |
|
not modelled |
84.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 99 | c3cu5B_ |
|
not modelled |
84.7 |
10 |
PDB header:transcription regulator
Chain: B: PDB Molecule:two component transcriptional regulator, arac family;
PDBTitle: crystal structure of a two component transcriptional regulator arac2 from clostridium phytofermentans isdg
|
| 100 | c1yadD_ |
|
not modelled |
84.5 |
15 |
PDB header:transcription
Chain: D: PDB Molecule:regulatory protein teni;
PDBTitle: structure of teni from bacillus subtilis
|
| 101 | c2nu8D_ |
|
not modelled |
84.1 |
23 |
PDB header:ligase
Chain: D: PDB Molecule:succinyl-coa ligase [adp-forming] subunit alpha;
PDBTitle: c123at mutant of e. coli succinyl-coa synthetase
|
| 102 | d1vc4a_ |
|
not modelled |
84.1 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 103 | c2zwmA_ |
|
not modelled |
83.8 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulatory protein yycf;
PDBTitle: crystal structure of yycf receiver domain from bacillus2 subtilis
|
| 104 | c3a0rB_ |
|
not modelled |
83.8 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:response regulator;
PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
|
| 105 | c2x41A_ |
|
not modelled |
83.7 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: structure of beta-glucosidase 3b from thermotoga neapolitana2 in complex with glucose
|
| 106 | c2p10D_ |
|
not modelled |
83.5 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:mll9387 protein;
PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
|
| 107 | c2d4aC_ |
|
not modelled |
83.4 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: structure of the malate dehydrogenase from aeropyrum pernix
|
| 108 | c1hyhA_ |
|
not modelled |
83.3 |
27 |
PDB header:oxidoreductase (choh(d)-nad+(a))
Chain: A: PDB Molecule:l-2-hydroxyisocaproate dehydrogenase;
PDBTitle: crystal structure of l-2-hydroxyisocaproate dehydrogenase from2 lactobacillus confusus at 2.2 angstroms resolution-an example of3 strong asymmetry between subunits
|
| 109 | c3b2nA_ |
|
not modelled |
83.2 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:uncharacterized protein q99uf4;
PDBTitle: crystal structure of dna-binding response regulator, luxr family, from2 staphylococcus aureus
|
| 110 | c2gjlA_ |
|
not modelled |
83.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein pa1024;
PDBTitle: crystal structure of 2-nitropropane dioxygenase
|
| 111 | c3tqvA_ |
|
not modelled |
82.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide pyrophosphorylase;
PDBTitle: structure of the nicotinate-nucleotide pyrophosphorylase from2 francisella tularensis.
|
| 112 | c1zn2A_ |
|
not modelled |
82.9 |
10 |
PDB header:transcription regulator
Chain: A: PDB Molecule:response regulatory protein;
PDBTitle: low resolution structure of response regulator styr
|
| 113 | c3cfyA_ |
|
not modelled |
82.8 |
8 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative luxo repressor protein;
PDBTitle: crystal structure of signal receiver domain of putative luxo2 repressor protein from vibrio parahaemolyticus
|
| 114 | c3d0oA_ |
|
not modelled |
82.8 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase 1;
PDBTitle: crystal structure of lactate dehydrogenase from2 staphylococcus aureus
|
| 115 | c3inpA_ |
|
not modelled |
82.7 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
| 116 | d2a9pa1 |
|
not modelled |
82.6 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
| 117 | d1znna1 |
|
not modelled |
82.5 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:PdxS-like |
| 118 | d1ytda1 |
|
not modelled |
82.4 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
| 119 | c3nepX_ |
|
not modelled |
82.2 |
16 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:malate dehydrogenase;
PDBTitle: 1.55a resolution structure of malate dehydrogenase from salinibacter2 ruber
|
| 120 | c3ilmD_ |
|
not modelled |
82.2 |
19 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:alr3790 protein;
PDBTitle: crystal structure of the alr3790 protein from anabaena sp. northeast2 structural genomics consortium target nsr437h
|