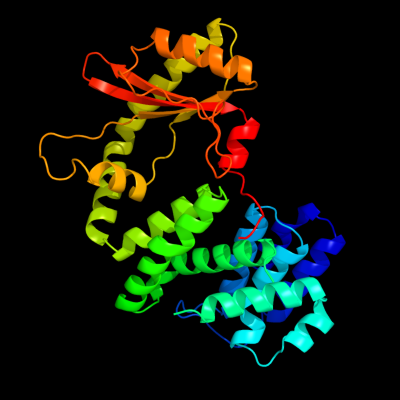
1 c1vj7B_
100.0
39
PDB header: hydrolase, transferaseChain: B: PDB Molecule: bifunctional rela/spot;PDBTitle: crystal structure of the bifunctional catalytic fragment of relseq,2 the rela/spot homolog from streptococcus equisimilis.
2 d1vj7a1
100.0
30
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain3 c3nr1A_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: hd domain-containing protein 3;PDBTitle: a metazoan ortholog of spot hydrolyzes ppgpp and plays a role in2 starvation responses
4 d1vj7a2
100.0
49
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: RelA/SpoT domain5 c3nqwB_
100.0
21
PDB header: hydrolaseChain: B: PDB Molecule: cg11900;PDBTitle: a metazoan ortholog of spot hydrolyzes ppgpp and plays a role in2 starvation responses
6 d2be3a1
100.0
21
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: RelA/SpoT domain7 c3l9dA_
99.9
24
PDB header: transferaseChain: A: PDB Molecule: putative gtp pyrophosphokinase;PDBTitle: the crystal structure of smu.1046c from streptococcus mutans ua159
8 c2kmmA_
99.9
49
PDB header: hydrolaseChain: A: PDB Molecule: guanosine-3',5'-bis(diphosphate) 3'-PDBTitle: solution nmr structure of the tgs domain of pg1808 from2 porphyromonas gingivalis. northeast structural genomics3 consortium target pgr122a (418-481)
9 c3hvzB_
99.9
59
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
10 c2ekiA_
99.8
32
PDB header: signaling proteinChain: A: PDB Molecule: developmentally-regulated gtp-binding protein 1;PDBTitle: solution structures of the tgs domain of human2 developmentally-regulated gtp-binding protein 1
11 d1tkea1
99.7
23
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: TGS domain12 d1wxqa2
99.7
35
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: G domain-linked domain13 d1nyra2
99.7
25
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: TGS domain14 c3ibwA_
99.6
23
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
15 c1wwtA_
99.5
18
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase, cytoplasmic;PDBTitle: solution structure of the tgs domain from human threonyl-2 trna synthetase
16 d1u8sa2
98.6
16
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor17 c1tkeA_
98.6
24
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase;PDBTitle: crystal structure of the editing domain of threonyl-trna2 synthetase complexed with serine
18 c2dwqB_
98.4
33
PDB header: hydrolaseChain: B: PDB Molecule: gtp-binding protein;PDBTitle: thermus thermophilus ychf gtp-binding protein
19 d1sc6a3
98.3
23
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain20 c2ohfA_
98.2
18
PDB header: hydrolaseChain: A: PDB Molecule: gtp-binding protein 9;PDBTitle: crystal structure of human ola1 in complex with amppcp
21 c1wxqA_
not modelled
98.2
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: gtp-binding protein;PDBTitle: crystal structure of gtp binding protein from pyrococcus horikoshii2 ot3
22 d1ygya3
not modelled
98.2
22
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain23 d2f1fa1
not modelled
98.2
16
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like24 d2fgca2
not modelled
98.1
9
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like25 d2pc6a2
not modelled
98.1
11
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like26 c1jalA_
not modelled
98.0
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ychf protein;PDBTitle: ychf protein (hi0393)
27 c2pc6C_
not modelled
98.0
11
PDB header: lyaseChain: C: PDB Molecule: probable acetolactate synthase isozyme iii (small subunit);PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
28 c2fgcA_
not modelled
97.9
10
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase, small subunit;PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
29 c1ni3A_
not modelled
97.9
24
PDB header: hydrolaseChain: A: PDB Molecule: ychf gtp-binding protein;PDBTitle: structure of the schizosaccharomyces pombe ychf gtpase
30 c2f1fA_
not modelled
97.9
16
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase isozyme iii small subunit;PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
31 c1nyqA_
not modelled
97.9
25
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase 1;PDBTitle: structure of staphylococcus aureus threonyl-trna synthetase2 complexed with an analogue of threonyl adenylate
32 c1qf6A_
not modelled
97.8
26
PDB header: ligase/rnaChain: A: PDB Molecule: threonyl-trna synthetase;PDBTitle: structure of e. coli threonyl-trna synthetase complexed with its2 cognate trna
33 d1zpva1
not modelled
97.6
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: SP0238-like34 c1u8sB_
not modelled
97.5
14
PDB header: transcriptionChain: B: PDB Molecule: glycine cleavage system transcriptionalPDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
35 c2nyiB_
97.5
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: unknown protein;PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
36 c3n0vD_
not modelled
97.5
12
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
37 c1y7pB_
not modelled
97.4
25
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein af1403;PDBTitle: 1.9 a crystal structure of a protein of unknown function2 af1403 from archaeoglobus fulgidus, probable metabolic3 regulator
38 d1u8sa1
not modelled
97.2
20
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor39 c3nrbD_
not modelled
97.1
17
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
40 c1ygyA_
not modelled
97.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
41 c1ybaC_
not modelled
97.0
24
PDB header: oxidoreductaseChain: C: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: the active form of phosphoglycerate dehydrogenase
42 c3o1lB_
not modelled
96.9
14
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
43 c3k5pA_
not modelled
96.9
19
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
44 c3obiC_
not modelled
96.6
20
PDB header: hydrolaseChain: C: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
45 c3louB_
not modelled
96.6
8
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
46 d2f06a2
not modelled
96.3
16
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like47 d1rwua_
not modelled
96.1
9
Fold: Ferredoxin-likeSuperfamily: YbeD/HP0495-likeFamily: YbeD-like48 c1rwuA_
not modelled
96.1
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical upf0250 protein ybed;PDBTitle: solution structure of conserved protein ybed from e. coli
49 d1phza1
not modelled
96.0
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain50 d1zud21
not modelled
95.7
23
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS51 c2qmxB_
not modelled
95.6
11
PDB header: ligaseChain: B: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of l-phe inhibited prephenate dehydratase from2 chlorobium tepidum tls
52 c2f06B_
not modelled
95.4
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
53 d2qmwa2
not modelled
95.3
17
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain54 d2hj1a1
not modelled
95.1
28
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: HI0395-like55 c2hj1A_
not modelled
95.1
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a 3d domain-swapped dimer of protein hi0395 from2 haemophilus influenzae
56 c2phmA_
not modelled
95.1
10
PDB header: oxidoreductaseChain: A: PDB Molecule: protein (phenylalanine-4-hydroxylase);PDBTitle: structure of phenylalanine hydroxylase dephosphorylated
57 c3mwbA_
not modelled
95.0
22
PDB header: lyaseChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
58 c3mtjA_
not modelled
94.7
21
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
59 d2f06a1
not modelled
94.5
20
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like60 c3cwiA_
not modelled
93.9
9
PDB header: biosynthetic proteinChain: A: PDB Molecule: thiamine-biosynthesis protein this;PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
61 c3luyA_
not modelled
93.8
14
PDB header: isomeraseChain: A: PDB Molecule: probable chorismate mutase;PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
62 d1tygb_
not modelled
93.8
19
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS63 c1tygG_
not modelled
92.2
19
PDB header: biosynthetic proteinChain: G: PDB Molecule: yjbs;PDBTitle: structure of the thiazole synthase/this complex
64 d1vjka_
not modelled
91.8
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: MoaD65 c2kl0A_
not modelled
91.8
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thiamin biosynthesis this;PDBTitle: solution nmr structure of rhodopseudomonas palustris rpa3574,2 northeast structural genomics consortium (nesg) target rpr325
66 c2qieB_
not modelled
91.6
14
PDB header: transferaseChain: B: PDB Molecule: molybdopterin synthase small subunit;PDBTitle: staphylococcus aureus molybdopterin synthase in complex2 with precursor z
67 c1tdjA_
not modelled
91.6
16
PDB header: allosteryChain: A: PDB Molecule: biosynthetic threonine deaminase;PDBTitle: threonine deaminase (biosynthetic) from e. coli
68 d2cu3a1
not modelled
91.4
18
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS69 c3po0A_
not modelled
90.8
16
PDB header: protein bindingChain: A: PDB Molecule: small archaeal modifier protein 1;PDBTitle: crystal structure of samp1 from haloferax volcanii
70 c2qmwA_
not modelled
90.4
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of the prephenate dehydratase (pdt) from2 staphylococcus aureus subsp. aureus mu50
71 c2dqbB_
not modelled
90.3
28
PDB header: hydrolase, dna binding proteinChain: B: PDB Molecule: deoxyguanosinetriphosphate triphosphohydrolase, putative;PDBTitle: crystal structure of dntp triphosphohydrolase from thermus2 thermophilus hb8, which is homologous to dgtp triphosphohydrolase
72 d1y7pa2
not modelled
90.0
25
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: AF1403 N-terminal domain-like73 c1dm9A_
not modelled
88.6
4
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical 15.5 kd protein in mrca-pckaPDBTitle: heat shock protein 15 kd
74 d1dm9a_
not modelled
88.6
4
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Heat shock protein 15 kD75 d2qgsa1
not modelled
88.5
14
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain76 c2ogiA_
not modelled
87.3
21
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein sag1661;PDBTitle: crystal structure of a putative metal dependent phosphohydrolase2 (sag1661) from streptococcus agalactiae serogroup v at 1.85 a3 resolution
77 c2dtjA_
not modelled
86.9
19
PDB header: transferaseChain: A: PDB Molecule: aspartokinase;PDBTitle: crystal structure of regulatory subunit of aspartate kinase2 from corynebacterium glutamicum
78 d1jala2
not modelled
86.2
31
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: G domain-linked domain79 c2o08B_
not modelled
86.2
19
PDB header: hydrolaseChain: B: PDB Molecule: bh1327 protein;PDBTitle: crystal structure of a putative hd superfamily hydrolase (bh1327) from2 bacillus halodurans at 1.90 a resolution
80 c3ccgA_
not modelled
85.4
14
PDB header: hydrolaseChain: A: PDB Molecule: hd superfamily hydrolase;PDBTitle: crystal structure of predicted hd superfamily hydrolase involved in2 nad metabolism (np_347894.1) from clostridium acetobutylicum at 1.503 a resolution
81 c2k6pA_
not modelled
84.8
18
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein hp_1423;PDBTitle: solution structure of hypothetical protein, hp1423
82 d2piea1
not modelled
84.5
19
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain83 d1fm0d_
not modelled
83.4
24
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: MoaD84 c3gqsB_
not modelled
82.0
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: adenylate cyclase-like protein;PDBTitle: crystal structure of the fha domain of ct664 protein from chlamydia2 trachomatis
85 d1ni3a2
not modelled
81.8
21
Fold: beta-Grasp (ubiquitin-like)Superfamily: TGS-likeFamily: G domain-linked domain86 d2cyya2
not modelled
80.5
19
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain87 c3fm8A_
not modelled
80.1
30
PDB header: transport protein/hydrolase activatorChain: A: PDB Molecule: kinesin-like protein kif13b;PDBTitle: crystal structure of full length centaurin alpha-1 bound with the fha2 domain of kif13b (capri target)
88 c3p96A_
not modelled
79.9
18
PDB header: hydrolaseChain: A: PDB Molecule: phosphoserine phosphatase serb;PDBTitle: crystal structure of phosphoserine phosphatase serb from mycobacterium2 avium, native form
89 d3djba1
not modelled
79.1
18
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain90 d1rwsa_
not modelled
79.0
35
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS91 d2joqa1
not modelled
77.7
9
Fold: Ferredoxin-likeSuperfamily: YbeD/HP0495-likeFamily: HP0495-like92 d3b57a1
not modelled
77.6
21
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain93 c3l76B_
not modelled
77.0
15
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of aspartate kinase from synechocystis
94 c2g1eA_
not modelled
76.7
13
PDB header: transferaseChain: A: PDB Molecule: hypothetical protein ta0895;PDBTitle: solution structure of ta0895
95 c2eh0A_
not modelled
76.2
26
PDB header: transport proteinChain: A: PDB Molecule: kinesin-like protein kif1b;PDBTitle: solution structure of the fha domain from human kinesin-2 like protein kif1b
96 d1c06a_
not modelled
75.1
23
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Ribosomal protein S497 d2g1la1
not modelled
75.0
22
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain98 c2bs2E_
not modelled
74.0
21
PDB header: oxidoreductaseChain: E: PDB Molecule: quinol-fumarate reductase iron-sulfur subunit b;PDBTitle: quinol:fumarate reductase from wolinella succinogenes
99 d2fug33
not modelled
73.3
22
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins100 d1yjma1
not modelled
72.9
21
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain101 c2re1A_
not modelled
72.9
14
PDB header: transferaseChain: A: PDB Molecule: aspartokinase, alpha and beta subunits;PDBTitle: crystal structure of aspartokinase alpha and beta subunits
102 c3bbnD_
not modelled
72.4
32
PDB header: ribosomeChain: D: PDB Molecule: ribosomal protein s4;PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
103 c2qjlA_
not modelled
72.2
26
PDB header: signaling proteinChain: A: PDB Molecule: ubiquitin-related modifier 1;PDBTitle: crystal structure of urm1
104 c3mzoA_
not modelled
71.8
7
PDB header: hydrolaseChain: A: PDB Molecule: lin2634 protein;PDBTitle: crystal structure of a hd-domain phosphohydrolase (lin2634) from2 listeria innocua at 1.98 a resolution
105 d1xo3a_
not modelled
71.3
36
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: C9orf74 homolog106 c2jqlA_
not modelled
71.0
35
PDB header: cell cycleChain: A: PDB Molecule: dna damage response protein kinase dun1;PDBTitle: nmr structure of the yeast dun1 fha domain in complex with2 a doubly phosphorylated (pt) peptide derived from rad533 scd1
107 d1gxca_
not modelled
70.7
17
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain108 c1gxcA_
not modelled
70.7
17
PDB header: phosphoprotein-binding domainChain: A: PDB Molecule: serine/threonine-protein kinase chk2;PDBTitle: fha domain from human chk2 kinase in complex with a2 synthetic phosphopeptide
109 d2uubd1
not modelled
70.5
32
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Ribosomal protein S4110 d2heka1
not modelled
70.5
24
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain111 d2pq7a1
not modelled
69.4
15
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain112 d1p9ka_
not modelled
69.2
14
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: YbcJ-like113 c3dwmA_
not modelled
68.9
15
PDB header: transferaseChain: A: PDB Molecule: 9.5 kda culture filtrate antigen cfp10a;PDBTitle: crystal structure of mycobacterium tuberculosis cyso, an antigen
114 d1g3ga_
not modelled
68.2
30
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain115 d1dmza_
not modelled
68.2
23
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain116 c3kt9A_
not modelled
68.0
17
PDB header: hydrolaseChain: A: PDB Molecule: aprataxin;PDBTitle: aprataxin fha domain
117 d3dtoa1
not modelled
68.0
20
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain118 d1vioa2
not modelled
67.5
36
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Pseudouridine synthase RsuA N-terminal domain119 d1wgka_
not modelled
67.2
25
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: C9orf74 homolog120 c3hx1B_
not modelled
67.1
36
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: slr1951 protein;PDBTitle: crystal structure of the slr1951 protein from synechocystis sp.2 northeast structural genomics consortium target sgr167a