1 c1zcdA_
99.9
14
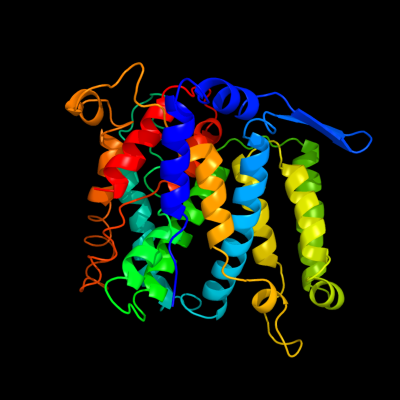
PDB header: membrane proteinChain: A: PDB Molecule: na(+)/h(+) antiporter 1;PDBTitle: crystal structure of the na+/h+ antiporter nhaa
2 c3jxoB_
99.3
30
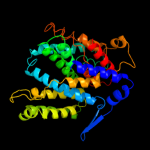
PDB header: transport proteinChain: B: PDB Molecule: trka-n domain protein;PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
3 d1vcta2
98.8
21
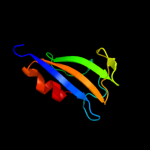
Fold: TrkA C-terminal domain-likeSuperfamily: TrkA C-terminal domain-likeFamily: TrkA C-terminal domain-like4 c2bknA_
98.8
18
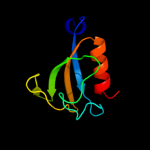
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical protein ph0236;PDBTitle: structure analysis of unknown function protein
5 d2fy8a2
98.8
18
Fold: TrkA C-terminal domain-likeSuperfamily: TrkA C-terminal domain-likeFamily: TrkA C-terminal domain-like6 c2fy8A_
98.5
18
PDB header: transport proteinChain: A: PDB Molecule: calcium-gated potassium channel mthk;PDBTitle: crystal structure of mthk rck domain in its ligand-free gating-ring2 form
7 c1lnqC_
98.5
18
PDB header: metal transportChain: C: PDB Molecule: potassium channel related protein;PDBTitle: crystal structure of mthk at 3.3 a
8 c3l4bG_
98.2
32
PDB header: transport proteinChain: G: PDB Molecule: trka k+ channel protien tm1088b;PDBTitle: crystal structure of an octomeric two-subunit trka k+ channel ring2 gating assembly, tm1088a:tm1088b, from thermotoga maritima
9 d3deda1
97.9
23
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like10 d2o1ra1
97.8
24
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like11 d2plia1
97.8
25
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like12 d2p13a1
97.7
18
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like13 c3llbA_
97.7
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of the protein pa3983 with unknown2 function from pseudomonas aeruginosa pao1
14 c3dedB_
97.6
25
PDB header: membrane proteinChain: B: PDB Molecule: probable hemolysin;PDBTitle: c-terminal domain of probable hemolysin from chromobacterium violaceum
15 d2o3ga1
97.6
23
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like16 d2plsa1
97.4
23
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like17 d2nqwa1
97.4
21
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like18 d2oaia1
97.3
18
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like19 d2rk5a1
97.2
15
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like20 d2r2za1
97.1
15
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like21 c3mt5A_
not modelled
96.8
11
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: potassium large conductance calcium-activated channel,PDBTitle: crystal structure of the human bk gating apparatus
22 d2p3ha1
not modelled
96.0
18
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like23 c3u6nC_
not modelled
95.4
10
PDB header: transport proteinChain: C: PDB Molecule: high-conductance ca2+-activated k+ channel protein;PDBTitle: open structure of the bk channel gating ring
24 d2p4pa1
not modelled
93.8
9
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like25 c3eywA_
not modelled
92.3
11
PDB header: transport proteinChain: A: PDB Molecule: c-terminal domain of glutathione-regulated potassium-effluxPDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
26 c2l0eA_
not modelled
88.8
27
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: structural and functional analysis of tm vi of the nhe1 isoform of the2 na+/h+ exchanger
27 d1lssa_
not modelled
88.2
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain28 c2g1uA_
not modelled
78.4
19
PDB header: transport proteinChain: A: PDB Molecule: hypothetical protein tm1088a;PDBTitle: crystal structure of a putative transport protein (tm1088a) from2 thermotoga maritima at 1.50 a resolution
29 c3llvA_
not modelled
78.2
16
PDB header: nad(p) binding proteinChain: A: PDB Molecule: exopolyphosphatase-related protein;PDBTitle: the crystal structure of the nad(p)-binding domain of an2 exopolyphosphatase-related protein from archaeoglobus fulgidus to3 1.7a
30 c3fwzA_
not modelled
74.4
10
PDB header: membrane proteinChain: A: PDB Molecule: inner membrane protein ybal;PDBTitle: crystal structure of trka-n domain of inner membrane protein ybal from2 escherichia coli
31 d1id1a_
not modelled
65.7
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain32 c2kbvA_
not modelled
53.0
35
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: structural and functional analysis of tm xi of the nhe12 isoform of the na+/h+ exchanger
33 c2khaA_
not modelled
48.2
12
PDB header: sugar binding proteinChain: A: PDB Molecule: beta-1,3-glucan-binding protein;PDBTitle: solution structure of a pathogen recognition domain from a2 lepidopteran insect, plodia interpunctella
34 c3ie4A_
not modelled
47.2
16
PDB header: immune systemChain: A: PDB Molecule: gram-negative binding protein 3;PDBTitle: b-glucan binding domain of drosophila gnbp3 defines a novel2 family of pattern recognition receptor
35 c2rqeA_
not modelled
44.9
12
PDB header: sugar binding proteinChain: A: PDB Molecule: beta-1,3-glucan-binding protein;PDBTitle: solution structure of the silkworm bgrp/gnbp3 n-terminal2 domain reveals the mechanism for b-1,3-glucan specific3 recognition
36 d2fy8a1
not modelled
43.5
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain37 d2c5aa1
not modelled
41.9
8
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases38 d1tygb_
not modelled
38.4
14
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS39 d1pjqa1
not modelled
36.2
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Siroheme synthase N-terminal domain-like40 c1tygG_
not modelled
35.6
15
PDB header: biosynthetic proteinChain: G: PDB Molecule: yjbs;PDBTitle: structure of the thiazole synthase/this complex
41 c3nafA_
not modelled
35.6
12
PDB header: ion transportChain: A: PDB Molecule: calcium-activated potassium channel subunit alpha-1;PDBTitle: structure of the intracellular gating ring from the human high-2 conductance ca2+ gated k+ channel (bk channel)
42 d2hmva1
not modelled
35.1
9
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Potassium channel NAD-binding domain43 c3dqpA_
not modelled
29.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase ylbe;PDBTitle: crystal structure of the oxidoreductase ylbe from2 lactococcus lactis, northeast structural genomics3 consortium target kr121.
44 d1hdoa_
not modelled
29.1
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases45 d1e5qa1
not modelled
28.5
4
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain46 c2kl0A_
not modelled
25.5
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative thiamin biosynthesis this;PDBTitle: solution nmr structure of rhodopseudomonas palustris rpa3574,2 northeast structural genomics consortium (nesg) target rpr325
47 c3cwiA_
not modelled
21.1
15
PDB header: biosynthetic proteinChain: A: PDB Molecule: thiamine-biosynthesis protein this;PDBTitle: crystal structure of thiamine biosynthesis protein (this)2 from geobacter metallireducens. northeast structural3 genomics consortium target gmr137
48 d1zud21
not modelled
19.5
21
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS49 c3c85A_
not modelled
19.2
9
PDB header: transport proteinChain: A: PDB Molecule: putative glutathione-regulated potassium-efflux systemPDBTitle: crystal structure of trka domain of putative glutathione-regulated2 potassium-efflux kefb from vibrio parahaemolyticus
50 d1b0aa1
not modelled
18.8
9
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain51 d1x5qa1
not modelled
18.4
14
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain52 d1edza1
not modelled
17.1
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain53 d1g9oa_
not modelled
17.0
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain54 c3r6dA_
not modelled
16.9
8
PDB header: lyase, isomeraseChain: A: PDB Molecule: nad-dependent epimerase/dehydratase;PDBTitle: crystal structure of nad-dependent epimerase/dehydratase from2 veillonella parvula dsm 2008 with cz-methylated lysine
55 c3bpuA_
not modelled
16.1
19
PDB header: transferaseChain: A: PDB Molecule: membrane-associated guanylate kinase, ww and pdz domain-PDBTitle: crystal structure of the 3rd pdz domain of human membrane associated2 guanylate kinase, c677s and c709s double mutant
56 d1g7sa2
not modelled
15.6
14
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors57 c3icpA_
not modelled
15.3
12
PDB header: isomeraseChain: A: PDB Molecule: nad-dependent epimerase/dehydratase;PDBTitle: crystal structure of udp-galactose 4-epimerase
58 c3l77A_
not modelled
15.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: short-chain alcohol dehydrogenase;PDBTitle: x-ray structure alcohol dehydrogenase from archaeon thermococcus2 sibiricus complexed with 5-hydroxy-nadp
59 c1edzA_
not modelled
15.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: 5,10-methylenetetrahydrofolate dehydrogenase;PDBTitle: structure of the nad-dependent 5,10-2 methylenetetrahydrofolate dehydrogenase from saccharomyces3 cerevisiae
60 d2cu3a1
not modelled
14.5
26
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: ThiS61 c3htrB_
not modelled
14.5
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized prc-barrel domain protein;PDBTitle: crystal structure of prc-barrel domain protein from2 rhodopseudomonas palustris
62 c3k8hA_
not modelled
14.2
14
PDB header: membrane proteinChain: A: PDB Molecule: 30klp;PDBTitle: structure of crystal form i of tp0453
63 c2vspA_
not modelled
14.1
15
PDB header: transport proteinChain: A: PDB Molecule: pdz domain-containing protein 1;PDBTitle: crystal structure of the fourth pdz domain of pdz domain-2 containing protein 1
64 c3r3sD_
not modelled
13.2
8
PDB header: oxidoreductaseChain: D: PDB Molecule: oxidoreductase;PDBTitle: structure of the ygha oxidoreductase from salmonella enterica
65 c2pk3B_
not modelled
13.2
13
PDB header: oxidoreductaseChain: B: PDB Molecule: gdp-6-deoxy-d-lyxo-4-hexulose reductase;PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
66 c2x4gA_
not modelled
13.1
9
PDB header: isomeraseChain: A: PDB Molecule: nucleoside-diphosphate-sugar epimerase;PDBTitle: crystal structure of pa4631, a nucleoside-diphosphate-sugar2 epimerase from pseudomonas aeruginosa
67 d1yjma1
not modelled
13.1
36
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain68 d1tp5a1
not modelled
13.1
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain69 c2q1wC_
not modelled
12.9
5
PDB header: sugar binding proteinChain: C: PDB Molecule: putative nucleotide sugar epimerase/ dehydratase;PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmh in2 complex with nad+
70 d2piea1
not modelled
12.8
19
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain71 d2i0ia1
not modelled
12.5
18
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain72 c1yj5C_
not modelled
12.3
38
PDB header: transferaseChain: C: PDB Molecule: 5' polynucleotide kinase-3' phosphatase fha domain;PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
73 d1a4ia1
not modelled
12.3
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain74 d1wvga1
not modelled
12.1
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases75 c3dhnA_
not modelled
11.9
19
PDB header: isomerase, lyaseChain: A: PDB Molecule: nad-dependent epimerase/dehydratase;PDBTitle: crystal structure of the putative epimerase q89z24_bactn2 from bacteroides thetaiotaomicron. northeast structural3 genomics consortium target btr310.
76 d2f1ka2
not modelled
11.8
10
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain77 c3ctmH_
not modelled
11.5
8
PDB header: oxidoreductaseChain: H: PDB Molecule: carbonyl reductase;PDBTitle: crystal structure of a carbonyl reductase from candida2 parapsilosis with anti-prelog stereo-specificity
78 c3h2sA_
not modelled
11.2
18
PDB header: oxidoreductaseChain: A: PDB Molecule: putative nadh-flavin reductase;PDBTitle: crystal structure of the q03b84 protein from lactobacillus2 casei. northeast structural genomics consortium target3 lcr19.
79 c2p5uC_
not modelled
11.0
8
PDB header: isomeraseChain: C: PDB Molecule: udp-glucose 4-epimerase;PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-2 epimerase complex with nad
80 c2nwqA_
not modelled
11.0
14
PDB header: oxidoreductaseChain: A: PDB Molecule: probable short-chain dehydrogenase;PDBTitle: short chain dehydrogenase from pseudomonas aeruginosa
81 d1rkxa_
not modelled
10.9
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases82 c3ew7A_
not modelled
10.8
9
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lmo0794 protein;PDBTitle: crystal structure of the lmo0794 protein from listeria2 monocytogenes. northeast structural genomics consortium3 target lmr162.
83 d1be9a_
not modelled
10.8
16
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain84 d1hi9a_
not modelled
10.6
13
Fold: Dipeptide transport proteinSuperfamily: Dipeptide transport proteinFamily: Dipeptide transport protein85 c2jxoA_
not modelled
10.5
18
PDB header: protein bindingChain: A: PDB Molecule: ezrin-radixin-moesin-binding phosphoprotein 50;PDBTitle: structure of the second pdz domain of nherf-1
86 d2brfa1
not modelled
10.4
32
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain87 d1dg6a_
not modelled
10.3
25
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like88 c1dg6A_
not modelled
10.3
25
PDB header: apoptosisChain: A: PDB Molecule: apo2l/tnf-related apopotis inducing ligand (trail);PDBTitle: crystal structure of apo2l/trail
89 c3edmD_
not modelled
10.3
8
PDB header: oxidoreductaseChain: D: PDB Molecule: short chain dehydrogenase;PDBTitle: crystal structure of a short chain dehydrogenase from agrobacterium2 tumefaciens
90 d2q8oa1
not modelled
10.3
20
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like91 c3kt9A_
not modelled
10.3
23
PDB header: hydrolaseChain: A: PDB Molecule: aprataxin;PDBTitle: aprataxin fha domain
92 c2qq5A_
not modelled
10.2
5
PDB header: oxidoreductaseChain: A: PDB Molecule: dehydrogenase/reductase sdr family member 1;PDBTitle: crystal structure of human sdr family member 1
93 d1xu9a_
not modelled
10.1
6
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases94 d2q1ma1
not modelled
9.9
26
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like95 d1lgpa_
not modelled
9.8
17
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain96 c2q30C_
not modelled
9.8
13
PDB header: unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a rmlc-like cupin protein (dde_2303) from2 desulfovibrio desulfuricans subsp. at 1.94 a resolution
97 c3iusB_
not modelled
9.6
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized conserved protein;PDBTitle: the structure of a functionally unknown conserved protein2 from silicibacter pomeroyi dss
98 c3oh8A_
not modelled
9.5
17
PDB header: isomeraseChain: A: PDB Molecule: nucleoside-diphosphate sugar epimerase (sula family);PDBTitle: crystal structure of the nucleoside-diphosphate sugar epimerase from2 corynebacterium glutamicum. northeast structural genomics consortium3 target cgr91
99 d1ujxa_
not modelled
9.5
41
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: FHA domain