1 c3nreB_
100.0
99
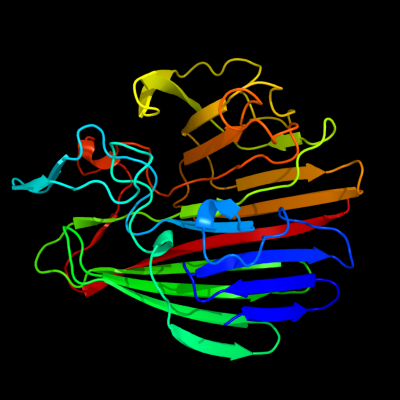
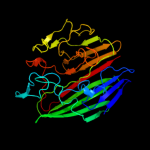
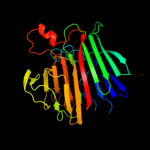
PDB header: isomeraseChain: B: PDB Molecule: aldose 1-epimerase;PDBTitle: crystal structure of a putative aldose 1-epimerase (b2544) from2 escherichia coli k12 at 1.59 a resolution
2 c3os7B_
100.0
19
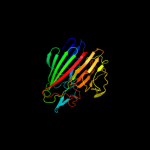
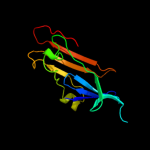
PDB header: isomeraseChain: B: PDB Molecule: galactose mutarotase-like protein;PDBTitle: crystal structure of a galactose mutarotase-like protein (ca_c0697)2 from clostridium acetobutylicum at 1.80 a resolution
3 c3os7D_
100.0
19
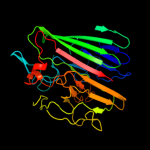
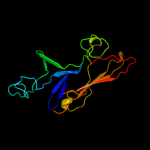
PDB header: isomeraseChain: D: PDB Molecule: galactose mutarotase-like protein;PDBTitle: crystal structure of a galactose mutarotase-like protein (ca_c0697)2 from clostridium acetobutylicum at 1.80 a resolution
4 c1ygaA_
100.0
15
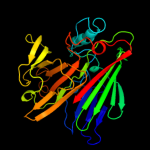
PDB header: isomeraseChain: A: PDB Molecule: hypothetical 37.9 kda protein in bio3-hxt17PDBTitle: crystal structure of saccharomyces cerevisiae yn9a protein,2 new york structural genomics consortium
5 d1z45a1
100.0
17
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)6 c3mwxA_
100.0
22
PDB header: isomeraseChain: A: PDB Molecule: aldose 1-epimerase;PDBTitle: crystal structure of a putative galactose mutarotase (bsu18360) from2 bacillus subtilis at 1.45 a resolution
7 c3imhB_
100.0
16
PDB header: isomeraseChain: B: PDB Molecule: galactose-1-epimerase;PDBTitle: crystal structure of galactose 1-epimerase from lactobacillus2 acidophilus ncfm
8 c3dcdA_
100.0
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: galactose mutarotase related enzyme;PDBTitle: x-ray structure of the galactose mutarotase related enzyme q5fkd7 from2 lactobacillus acidophilus at the resolution 1.9a. northeast3 structural genomics consortium target lar33.
9 c3q1nA_
100.0
17
PDB header: isomeraseChain: A: PDB Molecule: galactose mutarotase related enzyme;PDBTitle: crystal structure of a galactose mutarotase-like protein (lsei_2598)2 from lactobacillus casei atcc 334 at 1.61 a resolution
10 d1so0a_
100.0
20
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)11 d1lura_
100.0
15
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)12 d1nsza_
100.0
16
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)13 c3k25B_
100.0
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: slr1438 protein;PDBTitle: crystal structure of slr1438 protein from synechocystis sp. pcc 6803,2 northeast structural genomics consortium target sgr112
14 c1z45A_
100.0
17
PDB header: isomeraseChain: A: PDB Molecule: gal10 bifunctional protein;PDBTitle: crystal structure of the gal10 fusion protein galactose2 mutarotase/udp-galactose 4-epimerase from saccharomyces3 cerevisiae complexed with nad, udp-glucose, and galactose
15 c2htbB_
100.0
15
PDB header: isomeraseChain: B: PDB Molecule: putative enzyme related to aldose 1-epimerase;PDBTitle: crystal structure of a putative mutarotase (yead) from2 salmonella typhimurium in monoclinic form
16 c2cisA_
100.0
16
PDB header: isomeraseChain: A: PDB Molecule: hexose-6-phosphate mutarotase;PDBTitle: structure-based functional annotation: yeast ymr099c codes2 for a d-hexose-6-phosphate mutarotase. complex with3 tagatose-6-phosphate
17 d1jova_
100.0
20
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Hypothetical protein HI131718 c3ty1B_
100.0
13
PDB header: isomeraseChain: B: PDB Molecule: hypothetical aldose 1-epimerase;PDBTitle: crystal structure of a hypothetical aldose 1-epimerase (kpn_04629)2 from klebsiella pneumoniae subsp. pneumoniae mgh 78578 at 1.90 a3 resolution
19 c3bs6B_
97.1
16
PDB header: membrane protein, protein transportChain: B: PDB Molecule: inner membrane protein oxaa;PDBTitle: 1.8 angstrom crystal structure of the periplasmic domain of2 the membrane insertase yidc
20 c3blcB_
96.4
16
PDB header: chaperone,protein transportChain: B: PDB Molecule: inner membrane protein oxaa;PDBTitle: crystal structure of the periplasmic domain of the escherichia coli2 yidc
21 d1k1xa2
not modelled
89.8
12
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: 4-alpha-glucanotransferase, C-terminal domain22 c2xn1B_
not modelled
85.6
9
PDB header: hydrolaseChain: B: PDB Molecule: alpha-galactosidase;PDBTitle: structure of alpha-galactosidase from lactobacillus acidophilus ncfm2 with tris
23 c2cqtA_
not modelled
80.8
17
PDB header: transferaseChain: A: PDB Molecule: cellobiose phosphorylase;PDBTitle: crystal structure of cellvibrio gilvus cellobiose phosphorylase2 crystallized from sodium/potassium phosphate
24 c3rgbA_
not modelled
74.6
17
PDB header: oxidoreductaseChain: A: PDB Molecule: methane monooxygenase subunit b2;PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
25 c1yewI_
not modelled
74.6
17
PDB header: oxidoreductase, membrane proteinChain: I: PDB Molecule: particulate methane monooxygenase, b subunit;PDBTitle: crystal structure of particulate methane monooxygenase
26 c2yfnA_
not modelled
72.9
10
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase-sucrose kinase agask;PDBTitle: galactosidase domain of alpha-galactosidase-sucrose kinase,2 agask
27 d1ejxb_
not modelled
72.8
23
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit28 d4ubpb_
not modelled
68.5
23
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit29 c3mi6A_
not modelled
66.9
15
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase;PDBTitle: crystal structure of the alpha-galactosidase from lactobacillus2 brevis, northeast structural genomics consortium target lbr11.
30 c3rfrI_
not modelled
65.4
17
PDB header: oxidoreductaseChain: I: PDB Molecule: pmob;PDBTitle: crystal structure of particulate methane monooxygenase (pmmo) from2 methylocystis sp. strain m
31 d1jz8a4
not modelled
61.3
13
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: beta-Galactosidase, domain 532 d1e9ya1
not modelled
60.0
19
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit33 c3qgaD_
not modelled
57.7
32
PDB header: hydrolaseChain: D: PDB Molecule: fusion of urease beta and gamma subunits;PDBTitle: 3.0 a model of iron containing urease urea2b2 from helicobacter2 mustelae
34 c1v7wA_
not modelled
56.9
9
PDB header: transferaseChain: A: PDB Molecule: chitobiose phosphorylase;PDBTitle: crystal structure of vibrio proteolyticus chitobiose phosphorylase in2 complex with glcnac
35 d1v7wa2
not modelled
54.8
8
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Glycosyltransferase family 36 N-terminal domain36 c1e9zA_
not modelled
50.2
19
PDB header: hydrolaseChain: A: PDB Molecule: urease subunit alpha;PDBTitle: crystal structure of helicobacter pylori urease
37 c3isyA_
not modelled
45.5
15
PDB header: protein bindingChain: A: PDB Molecule: intracellular proteinase inhibitor;PDBTitle: crystal structure of an intracellular proteinase inhibitor (ipi,2 bsu11130) from bacillus subtilis at 2.61 a resolution
38 c2kl8A_
not modelled
40.2
13
PDB header: de novo proteinChain: A: PDB Molecule: or15;PDBTitle: solution nmr structure of de novo designed ferredoxin-like2 fold protein, northeast structural genomics consortium3 target or15
39 c3ndyG_
not modelled
40.1
19
PDB header: hydrolaseChain: G: PDB Molecule: endoglucanase d;PDBTitle: the structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
40 c1k1yA_
not modelled
36.9
10
PDB header: transferaseChain: A: PDB Molecule: 4-alpha-glucanotransferase;PDBTitle: crystal structure of thermococcus litoralis 4-alpha-glucanotransferase2 complexed with acarbose
41 c3qbtH_
not modelled
33.4
22
PDB header: protein transport/hydrolaseChain: H: PDB Molecule: inositol polyphosphate 5-phosphatase ocrl-1;PDBTitle: crystal structure of ocrl1 540-678 in complex with rab8a:gppnhp
42 c3bgaB_
not modelled
21.6
12
PDB header: hydrolaseChain: B: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of beta-galactosidase from bacteroides2 thetaiotaomicron vpi-5482
43 d2vzsa2
not modelled
20.9
37
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain44 c3obaA_
not modelled
18.6
15
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: structure of the beta-galactosidase from kluyveromyces lactis
45 d1w8oa1
not modelled
18.0
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes46 c2x3bB_
not modelled
12.2
27
PDB header: hydrolaseChain: B: PDB Molecule: toxic extracellular endopeptidase;PDBTitle: asap1 inactive mutant e294a, an extracellular toxic zinc2 metalloendopeptidase
47 d1f2ri_
not modelled
12.2
33
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain48 d2exna1
not modelled
10.8
12
Fold: MOSC N-terminal domain-likeSuperfamily: MOSC N-terminal domain-likeFamily: MOSC N-terminal domain-like49 d1c9fa_
not modelled
10.7
28
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain50 c2zf9D_
not modelled
10.4
31
PDB header: structural proteinChain: D: PDB Molecule: scae cell-surface anchored scaffoldin protein;PDBTitle: crystal structure of a type iii cohesin module from the cellulosomal2 scae cell-surface anchoring scaffoldin of ruminococcus flavefaciens
51 c2p28A_
not modelled
9.9
25
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-2;PDBTitle: structure of the phe2 and phe3 fragments of the integrin beta2 subunit
52 c2x41A_
not modelled
9.1
21
PDB header: hydrolaseChain: A: PDB Molecule: beta-glucosidase;PDBTitle: structure of beta-glucosidase 3b from thermotoga neapolitana2 in complex with glucose
53 c2kutA_
not modelled
8.4
37
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of gmr58a from geobacter metallireducens.2 northeast structural genomics consortium target gmr58a
54 d1ibxa_
not modelled
8.3
24
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain55 d1wn7a1
not modelled
8.2
11
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: DnaQ-like 3'-5' exonuclease56 c3qfgA_
not modelled
8.2
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: structure of a putative lipoprotein from staphylococcus aureus subsp.2 aureus nctc 8325
57 d1nkga3
not modelled
7.9
24
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Rhamnogalacturonase B, RhgB, N-terminal domain58 d1s58a_
not modelled
7.8
20
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: ssDNA virusesFamily: Parvoviridae-like VP59 c3nttA_
not modelled
7.6
28
PDB header: virusChain: A: PDB Molecule: capsid protein;PDBTitle: structural insights of adeno-associated virus 5. a gene therapy vector2 for cystic fibrosis
60 c3cfuA_
not modelled
7.6
17
PDB header: lipoproteinChain: A: PDB Molecule: uncharacterized lipoprotein yjha;PDBTitle: crystal structure of the yjha protein from bacillus2 subtilis. northeast structural genomics consortium target3 sr562
61 d2je8a2
not modelled
7.4
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain62 d1mspa_
not modelled
7.3
30
Fold: Immunoglobulin-like beta-sandwichSuperfamily: PapD-likeFamily: MSP-like63 d1ny721
not modelled
7.2
9
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP64 d1jv2a2
not modelled
7.1
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Integrin domainsFamily: Integrin domains65 c3ac0B_
not modelled
7.1
22
PDB header: hydrolaseChain: B: PDB Molecule: beta-glucosidase i;PDBTitle: crystal structure of beta-glucosidase from kluyveromyces marxianus in2 complex with glucose
66 d1oe1a1
not modelled
6.9
17
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins67 c1ibxB_
not modelled
6.6
29
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: chimera of igg binding protein g and dnaPDBTitle: nmr structure of dff40 and dff45 n-terminal domain complex
68 d1ibxb_
not modelled
6.6
29
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: CAD domain69 d1pgl21
not modelled
6.3
15
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP70 c3cdzA_
not modelled
6.1
31
PDB header: blood clottingChain: A: PDB Molecule: coagulation factor viii heavy chain;PDBTitle: crystal structure of human factor viii
71 d1exha_
not modelled
6.1
15
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Carbohydrate-binding domainFamily: Cellulose-binding domain family II72 d1pgw21
not modelled
6.1
15
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP73 c2r5kE_
not modelled
6.0
11
PDB header: viral proteinChain: E: PDB Molecule: major capsid protein l1;PDBTitle: pentamer structure of major capsid protein l1 of human2 papilloma virus type 11
74 c2zooA_
not modelled
5.9
10
PDB header: oxidoreductaseChain: A: PDB Molecule: probable nitrite reductase;PDBTitle: crystal structure of nitrite reductase from pseudoalteromonas2 haloplanktis tac125
75 c2l8aA_
not modelled
5.8
18
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: structure of a novel cbm3 lacking the calcium-binding site
76 c3qisA_
not modelled
5.4
25
PDB header: hydrolase/protein bindingChain: A: PDB Molecule: inositol polyphosphate 5-phosphatase ocrl-1;PDBTitle: recognition of the f&h motif by the lowe syndrome protein ocrl
77 c3mpbA_
not modelled
5.2
43
PDB header: isomeraseChain: A: PDB Molecule: sugar isomerase;PDBTitle: z5688 from e. coli o157:h7 bound to fructose
78 c2eelA_
not modelled
5.1
12
PDB header: apoptosisChain: A: PDB Molecule: cell death activator cide-a;PDBTitle: solution structure of the cide-n domain of human cell death2 activator cide-a
79 c2aenH_
not modelled
5.1
11
PDB header: viral proteinChain: H: PDB Molecule: outer capsid protein vp4, vp8* core;PDBTitle: crystal structure of the rotavirus strain ds-1 vp8* core
80 c1lf6A_
not modelled
5.0
10
PDB header: hydrolaseChain: A: PDB Molecule: glucoamylase;PDBTitle: crystal structure of bacterial glucoamylase