| 1 | c3mz1D_ |
|
 |
99.7 |
18 |
PDB header:transcription regulator
Chain: D: PDB Molecule:putative transcriptional regulator;
PDBTitle: the crystal structure of a possible transcription regulator protein2 from sinorhizobium meliloti 1021
|
| 2 | c3hhfB_ |
|
 |
99.7 |
20 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator, lysr family;
PDBTitle: structure of crga regulatory domain, a lysr-type transcriptional2 regulator from neisseria meningitidis.
|
| 3 | c3t1bB_ |
|
 |
99.5 |
18 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator, lysr family;
PDBTitle: crystal structure of the full-length aphb n100e variant
|
| 4 | c3hhgF_ |
|
 |
99.5 |
21 |
PDB header:transcription regulator
Chain: F: PDB Molecule:transcriptional regulator, lysr family;
PDBTitle: structure of crga, a lysr-type transcriptional regulator from2 neisseria meningitidis.
|
| 5 | c3fd3A_ |
|
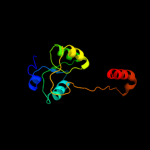 |
99.5 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:chromosome replication initiation inhibitor protein;
PDBTitle: structure of the c-terminal domains of a lysr family protein from2 agrobacterium tumefaciens str. c58.
|
| 6 | c3kosA_ |
|
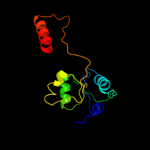 |
99.5 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:hth-type transcriptional activator ampr;
PDBTitle: structure of the ampr effector binding domain from citrobacter2 freundii
|
| 7 | c2uyeA_ |
|
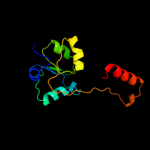 |
99.3 |
7 |
PDB header:transcription
Chain: A: PDB Molecule:regulatory protein;
PDBTitle: double mutant y110s,f111v dntr from burkholderia sp. strain2 dnt in complex with thiocyanate
|
| 8 | d1utha_ |
|
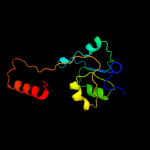 |
99.3 |
7 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 9 | c3oxnD_ |
|
 |
99.3 |
13 |
PDB header:transcription regulator
Chain: D: PDB Molecule:putative transcriptional regulator, lysr family;
PDBTitle: the crystal structure of a putative transcriptional regulator from2 vibrio parahaemolyticus
|
| 10 | c2qsxB_ |
|
 |
99.3 |
16 |
PDB header:transcription
Chain: B: PDB Molecule:putative transcriptional regulator, lysr family;
PDBTitle: crystal structure of putative transcriptional regulator lysr from2 vibrio parahaemolyticus
|
| 11 | c3ispA_ |
|
 |
99.2 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:hth-type transcriptional regulator
PDBTitle: crystal structure of argp from mycobacterium tuberculosis
|
| 12 | c2hxrA_ |
|
 |
99.2 |
11 |
PDB header:transcription
Chain: A: PDB Molecule:hth-type transcriptional regulator cynr;
PDBTitle: structure of the ligand binding domain of e. coli cynr, a2 transcriptional regulator controlling cyanate metabolism
|
| 13 | c1al3A_ |
|
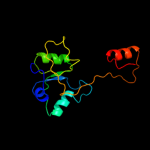 |
99.2 |
9 |
PDB header:transcription regulation
Chain: A: PDB Molecule:cys regulon transcriptional activator cysb;
PDBTitle: cofactor binding fragment of cysb from klebsiella aerogenes
|
| 14 | d1al3a_ |
|
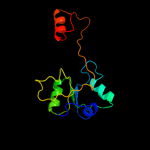 |
99.2 |
9 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 15 | c3fzvC_ |
|
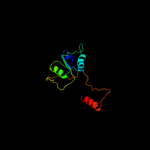 |
99.2 |
16 |
PDB header:transcription regulator
Chain: C: PDB Molecule:probable transcriptional regulator;
PDBTitle: crystal structure of pa01 protein, putative lysr family2 transcriptional regulator from pseudomonas aeruginosa
|
| 16 | c2ql3G_ |
|
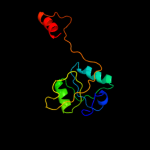 |
99.1 |
17 |
PDB header:transcription
Chain: G: PDB Molecule:probable transcriptional regulator, lysr family protein;
PDBTitle: crystal structure of the c-terminal domain of a probable lysr family2 transcriptional regulator from rhodococcus sp. rha1
|
| 17 | c3fzjC_ |
|
 |
99.1 |
10 |
PDB header:transcription regulator
Chain: C: PDB Molecule:lysr type regulator of tsambcd;
PDBTitle: tsar low resolution crystal structure, tetragonal form
|
| 18 | d1i6aa_ |
|
 |
99.1 |
12 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 19 | d2fyia1 |
|
 |
99.1 |
10 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 20 | c2h9qC_ |
|
 |
99.1 |
14 |
PDB header:transcription
Chain: C: PDB Molecule:hth-type transcriptional regulator catm;
PDBTitle: crystal structure of the effector binding domain of a catm2 variant (r156h)
|
| 21 | c1iz1B_ |
|
not modelled |
99.0 |
10 |
PDB header:dna binding protein
Chain: B: PDB Molecule:lysr-type regulatory protein;
PDBTitle: crystal structure of cbnr, a lysr family transcriptional2 regulator
|
| 22 | d2esna2 |
|
not modelled |
99.0 |
14 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 23 | c2esnC_ |
|
not modelled |
99.0 |
12 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:probable transcriptional regulator;
PDBTitle: the crystal structure of probable transcriptional regulator pa04772 from pseudomonas aeruginosa
|
| 24 | c3n6uA_ |
|
not modelled |
99.0 |
9 |
PDB header:transcription regulator
Chain: A: PDB Molecule:lysr type regulator of tsambcd;
PDBTitle: effector binding domain of tsar in complex with its inducer p-2 toluenesulfonate
|
| 25 | c3jv9B_ |
|
not modelled |
99.0 |
7 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator, lysr family;
PDBTitle: the structure of a reduced form of oxyr from n. meningitidis
|
| 26 | d1ixca2 |
|
not modelled |
98.9 |
9 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 27 | c3ho7A_ |
|
not modelled |
98.9 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:oxyr;
PDBTitle: crystal structure of oxyr from porphyromonas gingivalis
|
| 28 | c2f7cA_ |
|
not modelled |
98.8 |
13 |
PDB header:gene regulation
Chain: A: PDB Molecule:hth-type transcriptional regulator catm;
PDBTitle: catm effector binding domain with its effector cis,cis-muconate
|
| 29 | c2h9bB_ |
|
not modelled |
98.8 |
15 |
PDB header:transcription
Chain: B: PDB Molecule:hth-type transcriptional regulator benm;
PDBTitle: crystal structure of the effector binding domain of a benm variant2 (benm r156h/t157s)
|
| 30 | c2f78A_ |
|
not modelled |
98.6 |
15 |
PDB header:gene regulation
Chain: A: PDB Molecule:hth-type transcriptional regulator benm;
PDBTitle: benm effector binding domain with its effector benzoate
|
| 31 | c3onmB_ |
|
not modelled |
97.8 |
15 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator lrha;
PDBTitle: effector binding domain of lysr-type transcription factor rovm from y.2 pseudotuberculosis
|
| 32 | d1twya_ |
|
not modelled |
59.1 |
6 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 33 | c1twyG_ |
|
not modelled |
49.8 |
8 |
PDB header:structural genomics, unknown function
Chain: G: PDB Molecule:abc transporter, periplasmic substrate-binding protein;
PDBTitle: crystal structure of an abc-type phosphate transport receptor from2 vibrio cholerae
|
| 34 | c3c5tB_ |
|
not modelled |
36.4 |
27 |
PDB header:signaling protein/signaling protein
Chain: B: PDB Molecule:exendin-4;
PDBTitle: crystal structure of the ligand-bound glucagon-like peptide-1 receptor2 extracellular domain
|
| 35 | c3lr1A_ |
|
not modelled |
34.1 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:tungstate abc transporter, periplasmic tungstate-
PDBTitle: the crystal structure of the tungstate abc transporter from2 geobacter sulfurreducens
|
| 36 | c1vi7A_ |
|
not modelled |
33.4 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yigz;
PDBTitle: crystal structure of an hypothetical protein
|
| 37 | d1vi7a1 |
|
not modelled |
33.3 |
19 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:YigZ N-terminal domain-like |
| 38 | c3muqB_ |
|
not modelled |
31.6 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized conserved protein;
PDBTitle: the crystal structure of a conserved functionally unknown protein from2 vibrio parahaemolyticus rimd 2210633
|
| 39 | d1wj9a2 |
|
not modelled |
30.7 |
16 |
Fold:Ferredoxin-like
Superfamily:CRISPR-associated protein
Family:CRISPR-associated protein |
| 40 | d1amfa_ |
|
not modelled |
28.8 |
14 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 41 | c1wj9A_ |
|
not modelled |
25.6 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:crispr-associated protein;
PDBTitle: crystal structure of a crispr-associated protein from2 thermus thermophilus
|
| 42 | c3qrqA_ |
|
not modelled |
25.6 |
18 |
PDB header:rna binding protein/rna
Chain: A: PDB Molecule:putative uncharacterized protein tthb192;
PDBTitle: structure of thermus thermophilus cse3 bound to an rna representing a2 pre-cleavage complex
|
| 43 | d1atga_ |
|
not modelled |
19.4 |
4 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 44 | c2h5yC_ |
|
not modelled |
17.0 |
11 |
PDB header:metal transport
Chain: C: PDB Molecule:molybdate-binding periplasmic protein;
PDBTitle: crystallographic structure of the molybdate-binding protein of2 xanthomonas citri at 1.7 ang resolution bound to molybdate
|
| 45 | d2gk3a1 |
|
not modelled |
14.9 |
16 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:STM3548-like |
| 46 | c3rhtB_ |
|
not modelled |
14.2 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:(gatase1)-like protein;
PDBTitle: crystal structure of type 1 glutamine amidotransferase (gatase1)-like2 protein from planctomyces limnophilus
|
| 47 | d2ezwa1 |
|
not modelled |
12.3 |
18 |
Fold:Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
Superfamily:Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit
Family:Dimerization-anchoring domain of cAMP-dependent PK regulatory subunit |
| 48 | c2pksC_ |
|
not modelled |
11.6 |
13 |
PDB header:hydrolase/hydrolase inhibitor
Chain: C: PDB Molecule:thrombin heavy chain fragment;
PDBTitle: thrombin in complex with inhibitor
|
| 49 | d1sbpa_ |
|
not modelled |
10.1 |
7 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 50 | d1ezxc_ |
|
not modelled |
9.8 |
13 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |
| 51 | d1ixha_ |
|
not modelled |
8.4 |
10 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
| 52 | c1jrjA_ |
|
not modelled |
7.3 |
27 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:exendin-4;
PDBTitle: solution structure of exendin-4 in 30-vol% trifluoroethanol
|
| 53 | c3fj7A_ |
|
not modelled |
6.7 |
8 |
PDB header:protein binding
Chain: A: PDB Molecule:major antigenic peptide peb3;
PDBTitle: crystal structure of l-phospholactate bound peb3
|
| 54 | c1nauA_ |
|
not modelled |
6.6 |
0 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:glucagon;
PDBTitle: nmr solution structure of the glucagon antagonist [deshis1,2 desphe6, glu9]glucagon amide in the presence of3 perdeuterated dodecylphosphocholine micelles
|
| 55 | d1rgig3 |
|
not modelled |
6.4 |
10 |
Fold:Gelsolin-like
Superfamily:Actin depolymerizing proteins
Family:Gelsolin-like |
| 56 | d1iaua_ |
|
not modelled |
6.2 |
13 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Eukaryotic proteases |
| 57 | c1d0rA_ |
|
not modelled |
6.1 |
9 |
PDB header:hormone/growth factor
Chain: A: PDB Molecule:glucagon-like peptide-1-(7-36)-amide;
PDBTitle: solution structure of glucagon-like peptide-1-(7-36)-amide2 in trifluoroethanol/water
|
| 58 | d1j72a3 |
|
not modelled |
6.1 |
14 |
Fold:Gelsolin-like
Superfamily:Actin depolymerizing proteins
Family:Gelsolin-like |
| 59 | c1aksB_ |
|
not modelled |
6.0 |
7 |
PDB header:serine protease
Chain: B: PDB Molecule:alpha trypsin;
PDBTitle: crystal structure of the first active autolysate form of2 the porcine alpha trypsin
|
| 60 | d1os8a_ |
|
not modelled |
5.9 |
6 |
Fold:Trypsin-like serine proteases
Superfamily:Trypsin-like serine proteases
Family:Prokaryotic proteases |
| 61 | c1ca0C_ |
|
not modelled |
5.8 |
21 |
PDB header:complex (serine protease/inhibitor)
Chain: C: PDB Molecule:bovine chymotrypsin;
PDBTitle: bovine chymotrypsin complexed to appi
|
| 62 | c2cveA_ |
|
not modelled |
5.7 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ttha1053;
PDBTitle: crystal structure of a conserved hypothetical protein tt1547 from2 thermus thermophilus hb8
|
| 63 | d2qqra1 |
|
not modelled |
5.5 |
10 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:Tudor domain |
| 64 | c1lypA_ |
|
not modelled |
5.2 |
24 |
PDB header:lipopolysaccharide-binding protein
Chain: A: PDB Molecule:cap18;
PDBTitle: the solution structure of the active domain of cap18: a2 lipopolysaccharide binding protein from rabbit leukocytes
|
| 65 | c2mltB_ |
|
not modelled |
5.0 |
58 |
PDB header:toxin (hemolytic polypeptide)
Chain: B: PDB Molecule:melittin;
PDBTitle: melittin
|
| 66 | c1bh1A_ |
|
not modelled |
5.0 |
58 |
PDB header:toxin
Chain: A: PDB Molecule:melittin;
PDBTitle: structural studies of d-pro melittin, nmr, 20 structures
|

