1 c1y2iC_
100.0
97
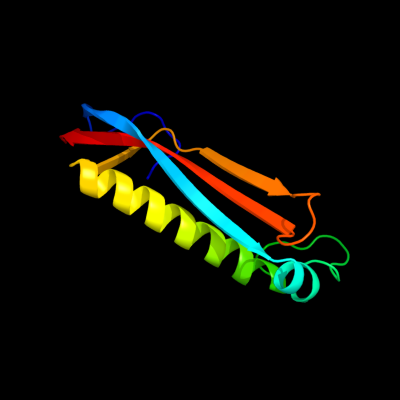
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein s0862;PDBTitle: crystal structure of mcsg target apc27401 from shigella2 flexneri
2 d1y2ia_
100.0
97
Fold: Dodecin subunit-likeSuperfamily: YbjQ-likeFamily: YbjQ-like3 d1vr4a1
100.0
55
Fold: Dodecin subunit-likeSuperfamily: YbjQ-likeFamily: YbjQ-like4 c3qkbB_
99.9
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function which belongs to2 pfam duf74 family (pepe_0654) from pediococcus pentosaceus atcc 257453 at 2.73 a resolution
5 c2y1bA_
97.7
19
PDB header: membrane proteinChain: A: PDB Molecule: putative outer membrane protein, signal;PDBTitle: crystal structure of the e. coli outer membrane lipoprotein2 rcsf
6 c2jz7A_
97.7
22
PDB header: selenium-binding proteinChain: A: PDB Molecule: selenium binding protein;PDBTitle: solution nmr structure of selenium-binding protein from2 methanococcus vannielii
7 c3bfjK_
23.9
12
PDB header: oxidoreductaseChain: K: PDB Molecule: 1,3-propanediol oxidoreductase;PDBTitle: crystal structure analysis of 1,3-propanediol oxidoreductase
8 c1xuzA_
18.0
33
PDB header: biosynthetic proteinChain: A: PDB Molecule: polysialic acid capsule biosynthesis protein siac;PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
9 c1vliA_
16.8
39
PDB header: biosynthetic proteinChain: A: PDB Molecule: spore coat polysaccharide biosynthesis protein spse;PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
10 c1zvaA_
16.6
24
PDB header: viral proteinChain: A: PDB Molecule: e2 glycoprotein;PDBTitle: a structure-based mechanism of sars virus membrane fusion
11 c2cx8B_
16.0
17
PDB header: transferaseChain: B: PDB Molecule: methyl transferase;PDBTitle: crystal structure of methyltransferase with ligand(sah)
12 d1vhka2
15.2
24
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: YggJ C-terminal domain-like13 d1gefa_
14.5
17
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hjc-like14 d1vlia2
13.8
39
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like15 d1hh1a_
13.5
24
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Hjc-like16 d1w96a2
13.1
25
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like17 c2qjhH_
12.8
46
PDB header: lyaseChain: H: PDB Molecule: putative aldolase mj0400;PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
18 c1z85B_
12.0
18
PDB header: transferaseChain: B: PDB Molecule: hypothetical protein tm1380;PDBTitle: crystal structure of a predicted rna methyltransferase (tm1380) from2 thermotoga maritima msb8 at 2.12 a resolution
19 d2j5ya1
12.0
40
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: GA module, an albumin-binding domain20 d1to3a_
11.9
25
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase21 c3gndC_
not modelled
11.9
31
PDB header: lyaseChain: C: PDB Molecule: aldolase lsrf;PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
22 c3jzdA_
not modelled
11.6
27
PDB header: oxidoreductaseChain: A: PDB Molecule: iron-containing alcohol dehydrogenase;PDBTitle: crystal structure of putative alcohol dehedrogenase (yp_298327.1) from2 ralstonia eutropha jmp134 at 2.10 a resolution
23 c2wj0B_
not modelled
11.6
32
PDB header: hydrolase/dnaChain: B: PDB Molecule: archaeal hjc;PDBTitle: crystal structures of holliday junction resolvases from2 archaeoglobus fulgidus bound to dna substrate
24 d1v6za2
not modelled
11.3
18
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: YggJ C-terminal domain-like25 d2zdra2
not modelled
10.8
28
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like26 c2v9dB_
not modelled
10.5
47
PDB header: lyaseChain: B: PDB Molecule: yage;PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
27 c3iv7B_
not modelled
10.4
19
PDB header: oxidoreductaseChain: B: PDB Molecule: alcohol dehydrogenase iv;PDBTitle: crystal structure of iron-containing alcohol dehydrogenase2 (np_602249.1) from corynebacterium glutamicum atcc 13032 kitasato at3 2.07 a resolution
28 d1a53a_
not modelled
10.2
46
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes29 c2rdcA_
not modelled
10.2
26
PDB header: lipid binding proteinChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative lipid binding protein (gsu0061) from2 geobacter sulfurreducens pca at 1.80 a resolution
30 c3cmgA_
not modelled
10.1
26
PDB header: hydrolaseChain: A: PDB Molecule: putative beta-galactosidase;PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
31 d2je8a5
not modelled
10.0
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases32 c3d0cB_
not modelled
9.8
53
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 oceanobacillus iheyensis at 1.9 a resolution
33 c3ox4D_
not modelled
9.5
19
PDB header: oxidoreductaseChain: D: PDB Molecule: alcohol dehydrogenase 2;PDBTitle: structures of iron-dependent alcohol dehydrogenase 2 from zymomonas2 mobilis zm4 complexed with nad cofactor
34 c2r94B_
not modelled
9.4
40
PDB header: lyaseChain: B: PDB Molecule: 2-keto-3-deoxy-(6-phospho-)gluconate aldolase;PDBTitle: crystal structure of kd(p)ga from t.tenax
35 d1ulza2
not modelled
9.3
31
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like36 c3mtjA_
not modelled
9.2
23
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
37 d1uf3a_
not modelled
9.0
37
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: TT1561-like38 c2egwB_
not modelled
8.6
23
PDB header: rna methyltransferaseChain: B: PDB Molecule: upf0088 protein aq_165;PDBTitle: crystal structure of rrna methyltransferase with sah ligand
39 c3sz8D_
not modelled
8.4
33
PDB header: transferaseChain: D: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase 2;PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
40 d1xxxa1
not modelled
8.3
27
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase41 c2nuxB_
not modelled
7.8
16
PDB header: lyaseChain: B: PDB Molecule: 2-keto-3-deoxygluconate/2-keto-3-deoxy-6-phospho gluconatePDBTitle: 2-keto-3-deoxygluconate aldolase from sulfolobus acidocaldarius,2 native structure in p6522 at 2.5 a resolution
42 d1rrma_
not modelled
7.6
24
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase43 d1vlja_
not modelled
7.5
24
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase44 c3jrkG_
not modelled
7.4
33
PDB header: lyaseChain: G: PDB Molecule: tagatose 1,6-diphosphate aldolase 2;PDBTitle: a putative tagatose 1,6-diphosphate aldolase from streptococcus2 pyogenes
45 c2rfgB_
not modelled
7.4
40
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
46 c2vc6A_
not modelled
7.4
33
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: structure of mosa from s. meliloti with pyruvate bound
47 d1o5ka_
not modelled
7.4
37
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase48 d1hl2a_
not modelled
7.3
40
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase49 d2j9ga2
not modelled
7.2
31
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like50 c2hdlA_
not modelled
7.2
24
PDB header: cytokineChain: A: PDB Molecule: small inducible cytokine b14;PDBTitle: solution structure of brak/cxcl14
51 c3stgA_
not modelled
7.2
20
PDB header: transferaseChain: A: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase;PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
52 d1p5dx4
not modelled
7.1
21
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain53 c3dz1A_
not modelled
7.1
26
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
54 c2ehhE_
not modelled
7.0
26
PDB header: lyaseChain: E: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 aquifex aeolicus
55 c3e96B_
not modelled
6.8
58
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 bacillus clausii
56 d2noca1
not modelled
6.8
31
Fold: Dodecin subunit-likeSuperfamily: YdgH-likeFamily: YdgH-like57 d2a6na1
not modelled
6.8
13
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase58 d1o2da_
not modelled
6.7
24
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase59 c3hl0B_
not modelled
6.7
32
PDB header: oxidoreductaseChain: B: PDB Molecule: maleylacetate reductase;PDBTitle: crystal structure of maleylacetate reductase from agrobacterium2 tumefaciens
60 c1vhkA_
not modelled
6.7
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yqeu;PDBTitle: crystal structure of an hypothetical protein
61 c3lerA_
not modelled
6.4
26
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from2 campylobacter jejuni subsp. jejuni nctc 11168
62 d1geqa_
not modelled
6.4
21
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes63 c2r8wB_
not modelled
6.3
37
PDB header: lyaseChain: B: PDB Molecule: agr_c_1641p;PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
64 c3lciA_
not modelled
6.2
32
PDB header: lyaseChain: A: PDB Molecule: n-acetylneuraminate lyase;PDBTitle: the d-sialic acid aldolase mutant v251w
65 c3g0sA_
not modelled
6.1
13
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: dihydrodipicolinate synthase from salmonella typhimurium lt2
66 d1nxza2
not modelled
6.1
14
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: YggJ C-terminal domain-like67 d1ejxb_
not modelled
6.0
57
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit68 c3fkkA_
not modelled
6.0
26
PDB header: lyaseChain: A: PDB Molecule: l-2-keto-3-deoxyarabonate dehydratase;PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
69 c3fluD_
not modelled
6.0
26
PDB header: lyaseChain: D: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
70 c3cprB_
not modelled
5.8
32
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthetase;PDBTitle: the crystal structure of corynebacterium glutamicum2 dihydrodipicolinate synthase to 2.2 a resolution
71 d2g7oa1
not modelled
5.8
21
Fold: TraM-likeSuperfamily: TraM-likeFamily: TraM-like72 c3b4uB_
not modelled
5.7
16
PDB header: lyaseChain: B: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihydrodipicolinate synthase from agrobacterium2 tumefaciens str. c58
73 d1w3ia_
not modelled
5.7
6
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase74 c3gjxD_
not modelled
5.6
22
PDB header: protein transportChain: D: PDB Molecule: exportin-1;PDBTitle: crystal structure of the nuclear export complex crm1-2 snurportin1-rangtp
75 c3n2xB_
not modelled
5.6
37
PDB header: lyaseChain: B: PDB Molecule: uncharacterized protein yage;PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
76 c3pueA_
not modelled
5.5
42
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
77 c2yxgD_
not modelled
5.4
42
PDB header: lyaseChain: D: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: crystal structure of dihyrodipicolinate synthase (dapa)
78 c2hmcA_
not modelled
5.4
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: the crystal structure of dihydrodipicolinate synthase dapa from2 agrobacterium tumefaciens
79 c3bi8A_
not modelled
5.4
21
PDB header: lyaseChain: A: PDB Molecule: dihydrodipicolinate synthase;PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
80 c3na8A_
not modelled
5.3
32
PDB header: lyaseChain: A: PDB Molecule: putative dihydrodipicolinate synthetase;PDBTitle: crystal structure of a putative dihydrodipicolinate synthetase from2 pseudomonas aeruginosa
81 d1x51a1
not modelled
5.3
17
Fold: NudixSuperfamily: NudixFamily: MutY C-terminal domain-like82 c2kxhB_
not modelled
5.2
29
PDB header: protein bindingChain: B: PDB Molecule: peptide of far upstream element-binding protein 1;PDBTitle: solution structure of the first two rrm domains of fir in the complex2 with fbp nbox peptide
83 d1tvga_
not modelled
5.0
25
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: APC10-like