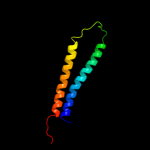
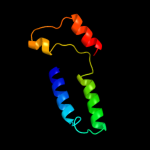
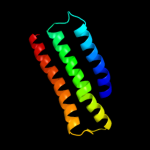
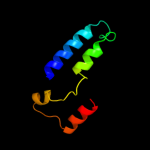
1 c2qfiB_
100.0
22
PDB header: transport proteinChain: B: PDB Molecule: ferrous-iron efflux pump fief;PDBTitle: structure of the zinc transporter yiip
2 d2qfia2
100.0
25
Fold: Cation efflux protein transmembrane domain-likeSuperfamily: Cation efflux protein transmembrane domain-likeFamily: Cation efflux protein transmembrane domain-like3 c2zztA_
99.3
22
PDB header: transport proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the cytosolic domain of the cation2 diffusion facilitator family protein
4 d3bypa1
99.3
19
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Cation efflux protein cytoplasmic domain-likeFamily: Cation efflux protein cytoplasmic domain-like5 d2qfia1
99.2
13
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Cation efflux protein cytoplasmic domain-likeFamily: Cation efflux protein cytoplasmic domain-like6 c2jsxA_
83.9
21
PDB header: chaperoneChain: A: PDB Molecule: protein napd;PDBTitle: solution structure of the e. coli tat proofreading2 chaperone protein napd
7 d2cu6a1
80.7
12
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like8 d1uwda_
68.4
8
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: PaaD-like9 c3lnoA_
49.3
11
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of domain of unknown function duf59 from2 bacillus anthracis
10 c2jp3A_
38.7
14
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
11 c2zxeG_
30.1
29
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: phospholemman-like protein;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
12 c3b9yA_
27.5
15
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter family rh-like protein;PDBTitle: crystal structure of the nitrosomonas europaea rh protein
13 c3pdsA_
25.0
9
PDB header: membrane protein/hydrolaseChain: A: PDB Molecule: fusion protein beta-2 adrenergic receptor/lysozyme;PDBTitle: irreversible agonist-beta2 adrenoceptor complex
14 c2kpsA_
22.6
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of domain iv from the ybbr family protein of2 desulfitobacterium hafniense
15 c2k2pA_
21.2
3
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1203;PDBTitle: solution nmr structure of protein atu1203 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att10, ontario center for structural proteomics target atc1183
16 c1v55B_
20.6
15
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: bovine heart cytochrome c oxidase at the fully reduced state
17 d1dlca3
20.4
12
Fold: Toxins' membrane translocation domainsSuperfamily: delta-Endotoxin (insectocide), N-terminal domainFamily: delta-Endotoxin (insectocide), N-terminal domain18 d1u19a_
19.8
15
Fold: Family A G protein-coupled receptor-likeSuperfamily: Family A G protein-coupled receptor-likeFamily: Rhodopsin-like19 d1ji6a3
19.8
17
Fold: Toxins' membrane translocation domainsSuperfamily: delta-Endotoxin (insectocide), N-terminal domainFamily: delta-Endotoxin (insectocide), N-terminal domain20 c1oy8A_
18.8
13
PDB header: membrane proteinChain: A: PDB Molecule: acriflavine resistance protein b;PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
21 c2ksaA_
not modelled
16.0
4
PDB header: neuropeptide receptor/neuropeptideChain: A: PDB Molecule: substance-p receptor;PDBTitle: substance p in dmpc/chaps isotropic q=0.25 bicelles as a ligand for2 nk1r
22 d1i1ga2
not modelled
16.0
11
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain23 d1ddfa_
not modelled
15.7
24
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD24 c2jo1A_
not modelled
15.4
35
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
25 c3rkoK_
not modelled
15.1
14
PDB header: oxidoreductaseChain: K: PDB Molecule: nadh-quinone oxidoreductase subunit k;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
26 c1ji6A_
not modelled
14.1
17
PDB header: toxinChain: A: PDB Molecule: pesticidial crystal protein cry3bb;PDBTitle: crystal structure of the insecticidal bacterial del2 endotoxin cry3bb1 bacillus thuringiensis
27 d1x38a2
not modelled
13.9
8
Fold: Flavodoxin-likeSuperfamily: Beta-D-glucan exohydrolase, C-terminal domainFamily: Beta-D-glucan exohydrolase, C-terminal domain28 c2vt4D_
not modelled
13.8
11
PDB header: receptorChain: D: PDB Molecule: beta1 adrenergic receptor;PDBTitle: turkey beta1 adrenergic receptor with stabilising mutations2 and bound cyanopindolol
29 c1u8sB_
not modelled
13.6
6
PDB header: transcriptionChain: B: PDB Molecule: glycine cleavage system transcriptionalPDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
30 c3mc8A_
not modelled
12.1
14
PDB header: membrane proteinChain: A: PDB Molecule: alr2269 protein;PDBTitle: potra1-3 of the periplasmic domain of omp85 from anabaena
31 c2ziyA_
not modelled
12.0
8
PDB header: signaling proteinChain: A: PDB Molecule: rhodopsin;PDBTitle: crystal structure of squid rhodopsin
32 d1fftb2
not modelled
11.9
10
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region33 c3eb7B_
not modelled
11.6
17
PDB header: toxinChain: B: PDB Molecule: insecticidal delta-endotoxin cry8ea1;PDBTitle: crystal structure of insecticidal delta-endotoxin cry8ea1 from2 bacillus thuringiensis at 2.2 angstroms resolution
34 c2djwF_
not modelled
11.6
21
PDB header: unknown functionChain: F: PDB Molecule: probable transcriptional regulator, asnc family;PDBTitle: crystal structure of ttha0845 from thermus thermophilus hb8
35 c2kj5A_
not modelled
11.4
40
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: phage integrase;PDBTitle: solution nmr structure of a domain from a putative phage2 integrase protein nmul_a0064 from nitrosospira multiformis,3 northeast structural genomics consortium target nmr46c
36 c3ibwA_
not modelled
10.9
13
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
37 c2crlA_
not modelled
10.6
17
PDB header: chaperoneChain: A: PDB Molecule: copper chaperone for superoxide dismutase;PDBTitle: the apo form of hma domain of copper chaperone for2 superoxide dismutase
38 c2zbcH_
not modelled
10.3
21
PDB header: transcriptionChain: H: PDB Molecule: 83aa long hypothetical transcriptional regulator asnc;PDBTitle: crystal structure of sts042, a stand-alone ram module protein, from2 hyperthermophilic archaeon sulfolobus tokodaii strain7.
39 c2rh1A_
not modelled
10.3
9
PDB header: membrane protein / hydrolaseChain: A: PDB Molecule: beta-2-adrenergic receptor/t4-lysozyme chimera;PDBTitle: high resolution crystal structure of human b2-adrenergic g protein-2 coupled receptor.
40 d2cg4a2
not modelled
10.3
13
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain41 d1iwga1
not modelled
9.1
13
Fold: Ferredoxin-likeSuperfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomainsFamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains42 c3rzeA_
not modelled
9.0
6
PDB header: hydrolaseChain: A: PDB Molecule: histamine h1 receptor, lysozyme chimera;PDBTitle: structure of the human histamine h1 receptor in complex with doxepin
43 c1ex1A_
not modelled
8.9
8
PDB header: hydrolaseChain: A: PDB Molecule: protein (beta-d-glucan exohydrolase isoenzyme exo1);PDBTitle: beta-d-glucan exohydrolase from barley
44 c2kt9A_
not modelled
8.7
35
PDB header: ribosomal proteinChain: A: PDB Molecule: probable 30s ribosomal protein psrp-3;PDBTitle: solution nmr structure of probable 30s ribosomal protein2 psrp-3 (ycf65-like protein) from synechocystis sp. (strain3 pcc 6803), northeast structural genomics consortium target4 target sgr46
45 c3emlA_
not modelled
8.5
23
PDB header: membrane protein, receptorChain: A: PDB Molecule: human adenosine a2a receptor/t4 lysozyme chimera;PDBTitle: the 2.6 a crystal structure of a human a2a adenosine2 receptor bound to zm241385.
46 c2e1aD_
not modelled
8.5
18
PDB header: transcriptionChain: D: PDB Molecule: 75aa long hypothetical regulatory protein asnc;PDBTitle: crystal structure of ffrp-dm1
47 c1dlcA_
not modelled
8.5
12
PDB header: toxinChain: A: PDB Molecule: delta-endotoxin cryiiia;PDBTitle: crystal structure of insecticidal delta-endotoxin from2 bacillus thuringiensis at 2.5 angstroms resolution
48 d1s6ua_
not modelled
8.4
3
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain49 c2aj1A_
not modelled
7.7
3
PDB header: hydrolaseChain: A: PDB Molecule: probable cadmium-transporting atpase;PDBTitle: solution structure of apocada
50 d1rl2a2
not modelled
7.7
28
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like51 d1ysja2
not modelled
7.7
9
Fold: Ferredoxin-likeSuperfamily: Bacterial exopeptidase dimerisation domainFamily: Bacterial exopeptidase dimerisation domain52 c2l3mA_
not modelled
7.6
7
PDB header: metal binding proteinChain: A: PDB Molecule: copper-ion-binding protein;PDBTitle: solution structure of the putative copper-ion-binding protein from2 bacillus anthracis str. ames
53 c2kppA_
not modelled
7.4
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin0431 protein;PDBTitle: solution nmr structure of lin0431 protein from listeria innocua.2 northeast structural genomics consortium target lkr112
54 c2k5eA_
not modelled
7.4
50
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195
55 c2yvxD_
not modelled
7.4
15
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
56 c3aynA_
not modelled
7.3
8
PDB header: signaling proteinChain: A: PDB Molecule: rhodopsin;PDBTitle: crystal structure of squid isorhodopsin
57 c2z73A_
not modelled
7.3
8
PDB header: membrane proteinChain: A: PDB Molecule: rhodopsin;PDBTitle: crystal structure of squid rhodopsin
58 c3aymA_
not modelled
7.3
8
PDB header: signaling proteinChain: A: PDB Molecule: rhodopsin;PDBTitle: crystal structure of the batho intermediate of squid rhodopsin
59 c2rogA_
not modelled
7.3
7
PDB header: metal binding proteinChain: A: PDB Molecule: heavy metal binding protein;PDBTitle: solution structure of thermus thermophilus hb8 ttha17182 protein in living e. coli cells
60 c2jxpA_
not modelled
7.1
38
PDB header: lipoproteinChain: A: PDB Molecule: putative lipoprotein b;PDBTitle: solution nmr structure of uncharacterized lipoprotein b2 from nitrosomonas europaea. northeast structural genomics3 target ner45a
61 d3ehbb2
not modelled
6.8
14
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region62 c2kt2A_
not modelled
6.7
10
PDB header: oxidoreductaseChain: A: PDB Molecule: mercuric reductase;PDBTitle: structure of nmera, the n-terminal hma domain of tn501 mercuric2 reductase
63 c3ac0B_
not modelled
6.7
13
PDB header: hydrolaseChain: B: PDB Molecule: beta-glucosidase i;PDBTitle: crystal structure of beta-glucosidase from kluyveromyces marxianus in2 complex with glucose
64 c3dxsX_
not modelled
6.7
13
PDB header: hydrolaseChain: X: PDB Molecule: copper-transporting atpase ran1;PDBTitle: crystal structure of a copper binding domain from hma7, a p-2 type atpase
65 c1i1gA_
not modelled
6.7
11
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator lrpa;PDBTitle: crystal structure of the lrp-like transcriptional regulator from the2 archaeon pyrococcus furiosus
66 c3pblB_
not modelled
6.6
17
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: d(3) dopamine receptor, lysozyme chimera;PDBTitle: structure of the human dopamine d3 receptor in complex with2 eticlopride
67 d1ln6a_
not modelled
6.6
15
Fold: Family A G protein-coupled receptor-likeSuperfamily: Family A G protein-coupled receptor-likeFamily: Rhodopsin-like68 d2qamc2
not modelled
6.4
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like69 c2ropA_
not modelled
6.4
9
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 2;PDBTitle: solution structure of domains 3 and 4 of human atp7b
70 c2b2hA_
not modelled
6.3
15
PDB header: transport proteinChain: A: PDB Molecule: ammonium transporter;PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
71 c3n23E_
not modelled
6.3
10
PDB header: hydrolaseChain: E: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the high affinity complex between ouabain and the2 e2p form of the sodium-potassium pump
72 c1yg0A_
not modelled
6.3
13
PDB header: metal transportChain: A: PDB Molecule: cop associated protein;PDBTitle: solution structure of apo-copp from helicobacter pylori
73 c2z73B_
not modelled
6.2
8
PDB header: membrane proteinChain: B: PDB Molecule: rhodopsin;PDBTitle: crystal structure of squid rhodopsin
74 c3aynB_
not modelled
6.2
8
PDB header: signaling proteinChain: B: PDB Molecule: rhodopsin;PDBTitle: crystal structure of squid isorhodopsin
75 d2qifa1
not modelled
6.1
3
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain76 c2l3bA_
not modelled
5.9
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein found in conjugate transposon;PDBTitle: solution nmr structure of the bt_0084 lipoprotein from bacteroides2 thetaiotaomicron, northeast structural genomics consortium target3 btr376
77 c3oe6A_
not modelled
5.9
10
PDB header: signaling protein,hydrolaseChain: A: PDB Molecule: c-x-c chemokine receptor type 4, lysozyme chimera;PDBTitle: crystal structure of the cxcr4 chemokine receptor in complex with a2 small molecule antagonist it1t in i222 spacegroup
78 d1cpza_
not modelled
5.9
7
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain79 c2jmrA_
not modelled
5.9
63
PDB header: cell adhesionChain: A: PDB Molecule: fimf;PDBTitle: nmr structure of the e. coli type 1 pilus subunit fimf
80 c3ftjA_
not modelled
5.8
10
PDB header: hydrolaseChain: A: PDB Molecule: macrolide export atp-binding/permease proteinPDBTitle: crystal structure of the periplasmic region of macb from2 actinobacillus actinomycetemcomitans
81 c2e7xA_
not modelled
5.8
16
PDB header: transcription regulatorChain: A: PDB Molecule: 150aa long hypothetical transcriptional regulator;PDBTitle: structure of the lrp/asnc like transcriptional regulator from2 sulfolobus tokodaii 7 complexed with its cognate ligand
82 c2ofhX_
not modelled
5.8
14
PDB header: hydrolase, membrane proteinChain: X: PDB Molecule: zinc-transporting atpase;PDBTitle: solution structure of the n-terminal domain of the zinc(ii) atpase2 ziaa in its apo form
83 c2kcqA_
not modelled
5.7
55
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: mov34/mpn/pad-1 family;PDBTitle: solution structure of protein sru_2040 from salinibacter2 ruber (strain dsm 13855) . northeast structural genomics3 consortium target srr106
84 d2hiya1
not modelled
5.7
11
Fold: SP0830-likeSuperfamily: SP0830-likeFamily: SP0830-like85 c2lc4A_
not modelled
5.6
50
PDB header: structural proteinChain: A: PDB Molecule: pilp protein;PDBTitle: solution structure of pilp from pseudomonas aeruginosa
86 c3sn6R_
not modelled
5.6
10
PDB header: signaling protein/hydrolaseChain: R: PDB Molecule: lysozyme, beta-2 adrenergic receptor;PDBTitle: crystal structure of the beta2 adrenergic receptor-gs protein complex
87 d1v54b2
not modelled
5.5
17
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region88 c2ga7A_
not modelled
5.4
7
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the copper(i) form of the third metal-2 binding domain of atp7a protein (menkes disease protein)
89 c2iqcA_
not modelled
5.4
18
PDB header: protein bindingChain: A: PDB Molecule: fanconi anemia group f protein;PDBTitle: crystal structure of human fancf protein that functions in2 the assembly of a dna damage signaling complex
90 c2zw2B_
not modelled
5.3
8
PDB header: ligaseChain: B: PDB Molecule: putative uncharacterized protein sts178;PDBTitle: crystal structure of formylglycinamide ribonucleotide amidotransferase2 iii from sulfolobus tokodaii (stpurs)
91 c2cg4B_
not modelled
5.3
13
PDB header: transcriptionChain: B: PDB Molecule: regulatory protein asnc;PDBTitle: structure of e.coli asnc
92 c2js4A_
not modelled
5.3
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0434 protein bb2007;PDBTitle: solution nmr structure of bordetella bronchiseptica protein2 bb2007. northeast structural genomics consortium target3 bor54
93 c2k49A_
not modelled
5.2
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0339 protein so_3888;PDBTitle: solution nmr structure of upf0339 protein so3888 from shewanella2 oneidensis. northeast structural genomics consortium target sor190
94 d1u8sa2
not modelled
5.2
4
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor95 d2cyya2
not modelled
5.2
13
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Lrp/AsnC-like transcriptional regulator C-terminal domain96 c3lq9B_
not modelled
5.2
12
PDB header: signaling proteinChain: B: PDB Molecule: dna-damage-inducible transcript 4 protein;PDBTitle: crystal strucure of human redd1, a hypoxia-induced regulator2 of mtor
97 c3g79A_
not modelled
5.1
70
PDB header: oxidoreductaseChain: A: PDB Molecule: ndp-n-acetyl-d-galactosaminuronic acid dehydrogenase;PDBTitle: crystal structure of ndp-n-acetyl-d-galactosaminuronic acid2 dehydrogenase from methanosarcina mazei go1