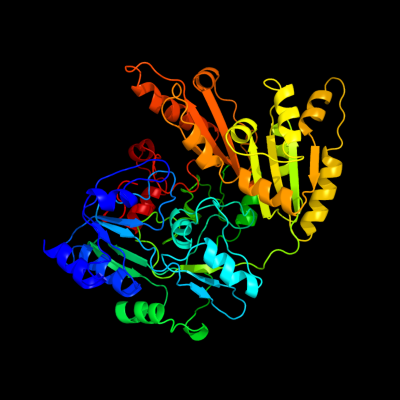
| 1 | c1f0xA_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-lactate dehydrogenase;
PDBTitle: crystal structure of d-lactate dehydrogenase, a peripheral2 membrane respiratory enzyme.
|
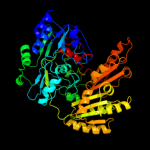
| 2 | c1ahuB_
|
|
 |
100.0 |
17 |
PDB header:flavoenzyme
Chain: B: PDB Molecule:vanillyl-alcohol oxidase;
PDBTitle: structure of the octameric flavoenzyme vanillyl-alcohol2 oxidase in complex with p-cresol
|
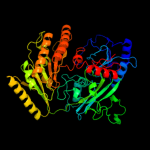
| 3 | c2uuvC_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:alkyldihydroxyacetonephosphate synthase;
PDBTitle: alkyldihydroxyacetonephosphate synthase in p1
|
| 4 | c3pm9A_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative dehydrogenase (rpa1076) from2 rhodopseudomonas palustris cga009 at 2.57 a resolution
|
| 5 | c1wveB_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-cresol dehydrogenase [hydroxylating]
PDBTitle: p-cresol methylhydroxylase: alteration of the structure of2 the flavoprotein subunit upon its binding to the3 cytochrome subunit
|
| 6 | c3bw7A_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytokinin dehydrogenase 1;
PDBTitle: maize cytokinin oxidase/dehydrogenase complexed with the allenic2 cytokinin analog ha-1
|
| 7 | c2exrA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytokinin dehydrogenase 7;
PDBTitle: x-ray structure of cytokinin oxidase/dehydrogenase (ckx)2 from arabidopsis thaliana at5g21482
|
| 8 | c2bvfA_
|
|
 |
100.0 |
22 |
PDB header:oxidase
Chain: A: PDB Molecule:6-hydroxy-d-nicotine oxidase;
PDBTitle: crystal structure of 6-hydoxy-d-nicotine oxidase from2 arthrobacter nicotinovorans. crystal form 3 (p1)
|
| 9 | c3fwaA_
|
|
 |
100.0 |
17 |
PDB header:flavoprotein
Chain: A: PDB Molecule:reticuline oxidase;
PDBTitle: structure of berberine bridge enzyme, c166a variant in complex with2 (s)-reticuline
|
| 10 | c3d2hA_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:berberine bridge-forming enzyme;
PDBTitle: structure of berberine bridge enzyme from eschscholzia californica,2 monoclinic crystal form
|
| 11 | c1zr6A_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucooligosaccharide oxidase;
PDBTitle: the crystal structure of an acremonium strictum glucooligosaccharide2 oxidase reveals a novel flavinylation
|
| 12 | c2ipiD_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:aclacinomycin oxidoreductase (aknox);
PDBTitle: crystal structure of aclacinomycin oxidoreductase
|
| 13 | c2wdwB_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative hexose oxidase;
PDBTitle: the native crystal structure of the primary hexose oxidase (2 dbv29) in antibiotic a40926 biosynthesis
|
| 14 | c3popD_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gilr oxidase;
PDBTitle: the crystal structure of gilr, an oxidoreductase that catalyzes the2 terminal step of gilvocarcin biosynthesis
|
| 15 | d1f0xa2
|
|
 |
100.0 |
20 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 16 | c2y3rC_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:taml;
PDBTitle: structure of the tirandamycin-bound fad-dependent2 tirandamycin oxidase taml in p21 space group
|
| 17 | c2vfvA_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xylitol oxidase;
PDBTitle: alditol oxidase from streptomyces coelicolor a3(2): complex2 with sulphite
|
| 18 | c1i19B_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cholesterol oxidase;
PDBTitle: crystal structure of cholesterol oxidase from b.sterolicum
|
| 19 | d1e8ga2
|
|
 |
100.0 |
22 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 20 | c3js8A_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cholesterol oxidase;
PDBTitle: solvent-stable cholesterol oxidase
|
| 21 | d1wvfa2 |
|
not modelled |
100.0 |
19 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 22 | d1w1oa2 |
|
not modelled |
100.0 |
24 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 23 | d2i0ka2 |
|
not modelled |
100.0 |
24 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:FAD-linked oxidases, N-terminal domain |
| 24 | d1f0xa1 |
|
not modelled |
100.0 |
15 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:D-lactate dehydrogenase |
| 25 | d1hska1 |
|
not modelled |
100.0 |
18 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase (MurB), N-terminal domain |
| 26 | d1uxya1 |
|
not modelled |
100.0 |
14 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase (MurB), N-terminal domain |
| 27 | d1e8ga1 |
|
not modelled |
100.0 |
14 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Vanillyl-alcohol oxidase-like |
| 28 | d1wvfa1 |
|
not modelled |
100.0 |
15 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Vanillyl-alcohol oxidase-like |
| 29 | c1hskA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: crystal structure of s. aureus murb
|
| 30 | c2yvsA_ |
|
not modelled |
99.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycolate oxidase subunit glce;
PDBTitle: crystal structure of glycolate oxidase subunit glce from thermus2 thermophilus hb8
|
| 31 | c1mbbA_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uridine diphospho-n-acetylenolpyruvylglucosamine
PDBTitle: oxidoreductase
|
| 32 | c3i99A_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvoylglucosamine reductase;
PDBTitle: the crystal structure of the udp-n-acetylenolpyruvoylglucosamine2 reductase from the vibrio cholerae o1 biovar tor
|
| 33 | c2gquA_ |
|
not modelled |
99.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:udp-n-acetylenolpyruvylglucosamine reductase;
PDBTitle: crystal structure of udp-n-acetylenolpyruvylglucosamine2 reductase (murb) from thermus caldophilus
|
| 34 | c3kwlA_
|
|
 |
99.6 |
14 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a hypothetical protein from helicobacter pylori
|
| 35 | d1nekb1
|
|
 |
99.4 |
25 |
Fold:Globin-like
Superfamily:alpha-helical ferredoxin
Family:Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain |
| 36 | c2h89B_
|
|
 |
99.3 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:succinate dehydrogenase ip subunit;
PDBTitle: avian respiratory complex ii with malonate bound
|
| 37 | c2b76N_
|
|
 |
99.2 |
17 |
PDB header:oxidoreductase
Chain: N: PDB Molecule:fumarate reductase iron-sulfur protein;
PDBTitle: e. coli quinol fumarate reductase frda e49q mutation
|
| 38 | c3cf4A_
|
|
 |
99.2 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:acetyl-coa decarboxylase/synthase alpha subunit;
PDBTitle: structure of the codh component of the m. barkeri acds complex
|
| 39 | c1nekB_
|
|
 |
99.2 |
24 |
PDB header:oxidoreductase/electron transport
Chain: B: PDB Molecule:succinate dehydrogenase iron-sulfur protein;
PDBTitle: complex ii (succinate dehydrogenase) from e. coli with2 ubiquinone bound
|
| 40 | d1kf6b1
|
|
 |
99.2 |
20 |
Fold:Globin-like
Superfamily:alpha-helical ferredoxin
Family:Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain |
| 41 | d2bs2b1 |
|
not modelled |
99.2 |
17 |
Fold:Globin-like
Superfamily:alpha-helical ferredoxin
Family:Fumarate reductase/Succinate dehydogenase iron-sulfur protein, C-terminal domain |
| 42 | c2bs2E_ |
|
not modelled |
99.1 |
16 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:quinol-fumarate reductase iron-sulfur subunit b;
PDBTitle: quinol:fumarate reductase from wolinella succinogenes
|
| 43 | d1w1oa1 |
|
not modelled |
98.7 |
17 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Cytokinin dehydrogenase 1 |
| 44 | c1gx7A_ |
|
not modelled |
98.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:periplasmic [fe] hydrogenase large subunit;
PDBTitle: best model of the electron transfer complex between2 cytochrome c3 and [fe]-hydrogenase
|
| 45 | c1c4cA_ |
|
not modelled |
98.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (fe-only hydrogenase);
PDBTitle: binding of exogenously added carbon monoxide at the active2 site of the fe-only hydrogenase (cpi) from clostridium3 pasteurianum
|
| 46 | c1hfeL_ |
|
not modelled |
98.0 |
21 |
PDB header:hydrogenase
Chain: L: PDB Molecule:protein (fe-only hydrogenase (e.c.1.18.99.1)
PDBTitle: 1.6 a resolution structure of the fe-only hydrogenase from2 desulfovibrio desulfuricans
|
| 47 | d1ffvc2 |
|
not modelled |
97.7 |
20 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 48 | c1ffuF_ |
|
not modelled |
97.6 |
19 |
PDB header:hydrolase
Chain: F: PDB Molecule:cutm, flavoprotein of carbon monoxide
PDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava which lacks the mo-pyranopterin moiety of the3 molybdenum cofactor
|
| 49 | c1n62C_ |
|
not modelled |
97.6 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carbon monoxide dehydrogenase medium chain;
PDBTitle: crystal structure of the mo,cu-co dehydrogenase (codh), n-2 butylisocyanide-bound state
|
| 50 | c2vdcI_ |
|
not modelled |
97.6 |
9 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:glutamate synthase [nadph] small chain;
PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
|
| 51 | c1t3qF_ |
|
not modelled |
97.5 |
22 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:quinoline 2-oxidoreductase medium subunit;
PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
|
| 52 | c3hrdC_ |
|
not modelled |
97.4 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nicotinate dehydrogenase fad-subunit;
PDBTitle: crystal structure of nicotinate dehydrogenase
|
| 53 | d1t3qc2 |
|
not modelled |
97.4 |
20 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 54 | d2c42a5 |
|
not modelled |
97.4 |
24 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 55 | d1v97a6 |
|
not modelled |
97.4 |
14 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 56 | d1n62c2 |
|
not modelled |
97.4 |
17 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 57 | c1gthD_ |
|
not modelled |
97.3 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydropyrimidine dehydrogenase;
PDBTitle: dihydropyrimidine dehydrogenase (dpd) from pig, ternary2 complex with nadph and 5-iodouracil
|
| 58 | d1gtea5 |
|
not modelled |
97.3 |
22 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 59 | d3b9jb2 |
|
not modelled |
97.3 |
14 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 60 | c3etrM_ |
|
not modelled |
97.2 |
11 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of xanthine oxidase in complex with2 lumazine
|
| 61 | c3b9jJ_ |
|
not modelled |
97.2 |
11 |
PDB header:oxidoreductase
Chain: J: PDB Molecule:xanthine oxidase;
PDBTitle: structure of xanthine oxidase with 2-hydroxy-6-methylpurine
|
| 62 | c2c3yA_ |
|
not modelled |
97.2 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate-ferredoxin oxidoreductase;
PDBTitle: crystal structure of the radical form of2 pyruvate:ferredoxin oxidoreductase from desulfovibrio3 africanus
|
| 63 | c2w3rG_ |
|
not modelled |
97.1 |
18 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:xanthine dehydrogenase;
PDBTitle: crystal structure of xanthine dehydrogenase (desulfo form)2 from rhodobacter capsulatus in complex with hypoxanthine
|
| 64 | d1jroa4 |
|
not modelled |
97.0 |
21 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 65 | c1rm6E_ |
|
not modelled |
96.9 |
24 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:4-hydroxybenzoyl-coa reductase beta subunit;
PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
|
| 66 | d3c8ya3 |
|
not modelled |
96.9 |
24 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 67 | c1wygA_ |
|
not modelled |
96.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of a rat xanthine dehydrogenase triple mutant2 (c535a, c992r and c1324s)
|
| 68 | d2fug91 |
|
not modelled |
96.7 |
31 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 69 | c2fugG_ |
|
not modelled |
96.7 |
31 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:nadh-quinone oxidoreductase chain 9;
PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
|
| 70 | d1hfel2 |
|
not modelled |
96.6 |
21 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 71 | d1rm6b2 |
|
not modelled |
96.6 |
25 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CO dehydrogenase flavoprotein N-terminal domain-like |
| 72 | d2fug34 |
|
not modelled |
96.5 |
12 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 73 | d1jb0c_ |
|
not modelled |
96.4 |
30 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:7-Fe ferredoxin |
| 74 | c2gmhA_ |
|
not modelled |
96.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:electron transfer flavoprotein-ubiquinone
PDBTitle: structure of porcine electron transfer flavoprotein-2 ubiquinone oxidoreductase in complexed with ubiquinone
|
| 75 | d1xera_ |
|
not modelled |
96.3 |
34 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Archaeal ferredoxins |
| 76 | d7fd1a_ |
|
not modelled |
96.2 |
24 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:7-Fe ferredoxin |
| 77 | d1clfa_ |
|
not modelled |
95.9 |
38 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Short-chain ferredoxins |
| 78 | d2fdna_ |
|
not modelled |
95.9 |
38 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Short-chain ferredoxins |
| 79 | c1kqfB_ |
|
not modelled |
95.9 |
34 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:formate dehydrogenase, nitrate-inducible, iron-sulfur
PDBTitle: formate dehydrogenase n from e. coli
|
| 80 | d1y5ib1 |
|
not modelled |
95.8 |
37 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 81 | d1bc6a_ |
|
not modelled |
95.7 |
31 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:7-Fe ferredoxin |
| 82 | d1fcaa_ |
|
not modelled |
95.4 |
40 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Short-chain ferredoxins |
| 83 | d1blua_ |
|
not modelled |
95.4 |
33 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Short-chain ferredoxins |
| 84 | d1rgva_ |
|
not modelled |
95.3 |
27 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Short-chain ferredoxins |
| 85 | c2fugC_ |
|
not modelled |
95.2 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh-quinone oxidoreductase chain 3;
PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
|
| 86 | d1dura_ |
|
not modelled |
95.2 |
39 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Short-chain ferredoxins |
| 87 | c2zvsB_ |
|
not modelled |
95.2 |
36 |
PDB header:electron transport
Chain: B: PDB Molecule:uncharacterized ferredoxin-like protein yfhl;
PDBTitle: crystal structure of the 2[4fe-4s] ferredoxin from escherichia coli
|
| 88 | c2ivfB_ |
|
not modelled |
95.2 |
37 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ethylbenzene dehydrogenase beta-subunit;
PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
|
| 89 | d1h98a_ |
|
not modelled |
95.1 |
31 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:7-Fe ferredoxin |
| 90 | c2fgoA_ |
|
not modelled |
95.1 |
29 |
PDB header:electron transport
Chain: A: PDB Molecule:ferredoxin;
PDBTitle: structure of the 2[4fe-4s] ferredoxin from pseudomonas2 aeruginosa
|
| 91 | d1jnrb_ |
|
not modelled |
94.5 |
26 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 92 | c2v2kB_ |
|
not modelled |
94.5 |
27 |
PDB header:electron transport
Chain: B: PDB Molecule:ferredoxin;
PDBTitle: the crystal structure of fdxa, a 7fe ferredoxin from2 mycobacterium smegmatis
|
| 93 | d1gtea1 |
|
not modelled |
94.5 |
18 |
Fold:Globin-like
Superfamily:alpha-helical ferredoxin
Family:Dihydropyrimidine dehydrogenase, N-terminal domain |
| 94 | c3gyxJ_ |
|
not modelled |
93.8 |
29 |
PDB header:oxidoreductase
Chain: J: PDB Molecule:adenylylsulfate reductase;
PDBTitle: crystal structure of adenylylsulfate reductase from2 desulfovibrio gigas
|
| 95 | c3c7bE_ |
|
not modelled |
93.1 |
25 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:sulfite reductase, dissimilatory-type subunit beta;
PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
|
| 96 | c2v4jE_ |
|
not modelled |
93.1 |
43 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
|
| 97 | d2gmha3 |
|
not modelled |
93.1 |
23 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:ETF-QO domain-like |
| 98 | c2vpyB_ |
|
not modelled |
92.9 |
47 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nrfc protein;
PDBTitle: polysulfide reductase with bound quinone inhibitor,2 pentachlorophenol (pcp)
|
| 99 | d1kqfb1 |
|
not modelled |
92.6 |
41 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 100 | d1h0hb_ |
|
not modelled |
92.6 |
24 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 101 | c1ti2F_ |
|
not modelled |
92.2 |
30 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:pyrogallol hydroxytransferase small subunit;
PDBTitle: crystal structure of pyrogallol-phloroglucinol2 transhydroxylase from pelobacter acidigallici
|
| 102 | d1iqza_ |
|
not modelled |
91.9 |
21 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Single 4Fe-4S cluster ferredoxin |
| 103 | c3c7bA_ |
|
not modelled |
91.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit alpha;
PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
|
| 104 | c1dwlA_ |
|
not modelled |
91.5 |
28 |
PDB header:electron transfer
Chain: A: PDB Molecule:ferredoxin i;
PDBTitle: the ferredoxin-cytochrome complex using heteronuclear nmr2 and docking simulation
|
| 105 | d1vjwa_ |
|
not modelled |
88.3 |
38 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Single 4Fe-4S cluster ferredoxin |
| 106 | d2i0ka1 |
|
not modelled |
87.3 |
15 |
Fold:Ferredoxin-like
Superfamily:FAD-linked oxidases, C-terminal domain
Family:Cholesterol oxidase |
| 107 | d1sj1a_ |
|
not modelled |
86.2 |
20 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Single 4Fe-4S cluster ferredoxin |
| 108 | c2v4jA_ |
|
not modelled |
85.2 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite reductase, dissimilatory-type subunit
PDBTitle: the crystal structure of desulfovibrio vulgaris2 dissimilatory sulfite reductase bound to dsrc provides3 novel insights into the mechanism of sulfate respiration
|
| 109 | d3c7bb1 |
|
not modelled |
85.0 |
25 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 110 | d1fxra_ |
|
not modelled |
84.9 |
19 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Single 4Fe-4S cluster ferredoxin |
| 111 | c2fugA_ |
|
not modelled |
84.0 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh-quinone oxidoreductase chain 1;
PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
|
| 112 | d1vlfn2 |
|
not modelled |
83.8 |
18 |
Fold:Ferredoxin-like
Superfamily:4Fe-4S ferredoxins
Family:Ferredoxin domains from multidomain proteins |
| 113 | c3hlyA_ |
|
not modelled |
77.3 |
11 |
PDB header:flavoprotein
Chain: A: PDB Molecule:flavodoxin-like domain;
PDBTitle: crystal structure of the flavodoxin-like domain from2 synechococcus sp q5mzp6_synp6 protein. northeast structural3 genomics consortium target snr135d.
|
| 114 | c3bk7A_ |
|
not modelled |
76.9 |
33 |
PDB header:hydrolyase/translation
Chain: A: PDB Molecule:abc transporter atp-binding protein;
PDBTitle: structure of the complete abce1/rnaase-l inhibitor protein2 from pyrococcus abysii
|
| 115 | d1jfla1 |
|
not modelled |
76.4 |
2 |
Fold:ATC-like
Superfamily:Aspartate/glutamate racemase
Family:Aspartate/glutamate racemase |
| 116 | c2uvaI_ |
|
not modelled |
75.5 |
19 |
PDB header:transferase
Chain: I: PDB Molecule:fatty acid synthase beta subunits;
PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the beta subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
|
| 117 | c2vkzH_ |
|
not modelled |
74.4 |
22 |
PDB header:transferase
Chain: H: PDB Molecule:fatty acid synthase subunit beta;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid2 synthase type i multienzyme complex
|
| 118 | d2plsa1 |
|
not modelled |
67.8 |
23 |
Fold:FAD-binding/transporter-associated domain-like
Superfamily:FAD-binding/transporter-associated domain-like
Family:CorC/HlyC domain-like |
| 119 | c2vdcF_ |
|
not modelled |
60.2 |
27 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glutamate synthase [nadph] large chain;
PDBTitle: the 9.5 a resolution structure of glutamate synthase from2 cryo-electron microscopy and its oligomerization behavior3 in solution: functional implications.
|
| 120 | c3fniA_ |
|
not modelled |
58.1 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative diflavin flavoprotein a 3;
PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
|