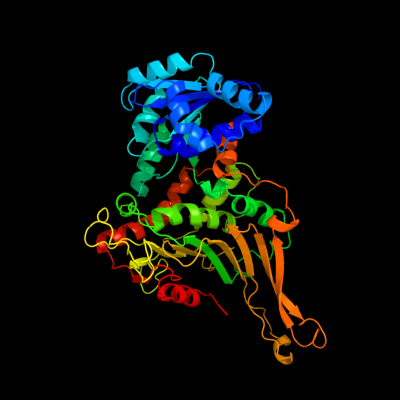
| 1 | c2bhlB_
|
|
 |
100.0 |
37 |
PDB header:oxidoreductase (choh(d)-nadp)
Chain: B: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray structure of human glucose-6-phosphate dehydrogenase2 (deletion variant) complexed with glucose-6-phosphate
|
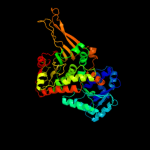
| 2 | c1h9aA_
|
|
 |
100.0 |
33 |
PDB header:oxidoreductase (choh(d) - nad(p))
Chain: A: PDB Molecule:glucose 6-phosphate 1-dehydrogenase;
PDBTitle: complex of active mutant (q365->c) of glucose 6-phosphate2 dehydrogenase from l. mesenteroides with coenzyme nadp
|
| 3 | c1qkiE_
|
|
 |
100.0 |
36 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray structure of human glucose 6-phosphate dehydrogenase2 (variant canton r459l) complexed with structural nadp+
|
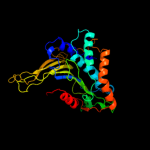
| 4 | d1qkia2
|
|
 |
100.0 |
41 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:Glucose 6-phosphate dehydrogenase-like |
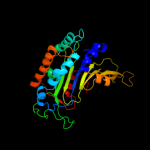
| 5 | d1h9aa2
|
|
 |
100.0 |
34 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:Glucose 6-phosphate dehydrogenase-like |
| 6 | d1h9aa1
|
|
 |
100.0 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 7 | d1qkia1
|
|
 |
100.0 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 8 | c3m2tA_
|
|
 |
97.8 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of dehydrogenase from chromobacterium2 violaceum
|
| 9 | c3btuD_
|
|
 |
97.8 |
20 |
PDB header:transcription
Chain: D: PDB Molecule:galactose/lactose metabolism regulatory protein
PDBTitle: crystal structure of the super-repressor mutant of gal80p2 from saccharomyces cerevisiae; gal80(s2) [e351k]
|
| 10 | d2nvwa1
|
|
 |
97.7 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 11 | c3kuxA_
|
|
 |
97.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of the ypo2259 putative oxidoreductase from yersinia pestis
|
| 12 | c3e18A_
|
|
 |
97.6 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of nad-binding protein from listeria innocua
|
| 13 | c3ezyB_
|
|
 |
97.4 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of probable dehydrogenase tm_0414 from2 thermotoga maritima
|
| 14 | c3db2C_
|
|
 |
97.4 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
| 15 | c1zh8B_
|
|
 |
97.3 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
| 16 | c3gfgB_
|
|
 |
97.3 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uncharacterized oxidoreductase yvaa;
PDBTitle: structure of putative oxidoreductase yvaa from bacillus subtilis in2 triclinic form
|
| 17 | d1h6da1
|
|
 |
97.3 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 18 | c3euwB_
|
|
 |
97.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|
| 19 | c1evjC_
|
|
 |
97.2 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: crystal structure of glucose-fructose oxidoreductase (gfor)2 delta1-22 s64d
|
| 20 | c2nvwB_
|
|
 |
97.1 |
14 |
PDB header:transcription
Chain: B: PDB Molecule:galactose/lactose metabolism regulatory protein
PDBTitle: crystal sctucture of transcriptional regulator gal80p from2 kluyveromymes lactis
|
| 21 | c3ec7C_ |
|
not modelled |
97.0 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dehydrogenase;
PDBTitle: crystal structure of putative dehydrogenase from salmonella2 typhimurium lt2
|
| 22 | c3e82A_ |
|
not modelled |
97.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from2 klebsiella pneumoniae
|
| 23 | c3uuwB_ |
|
not modelled |
97.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase with nad(p)-binding rossmann-fold
PDBTitle: 1.63 angstrom resolution crystal structure of dehydrogenase (mvim)2 from clostridium difficile.
|
| 24 | c2glxD_ |
|
not modelled |
96.9 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1,5-anhydro-d-fructose reductase;
PDBTitle: crystal structure analysis of bacterial 1,5-af reductase
|
| 25 | c3c1aB_ |
|
not modelled |
96.8 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase (zp_00056571.1) from2 magnetospirillum magnetotacticum ms-1 at 1.85 a resolution
|
| 26 | c3fd8A_ |
|
not modelled |
96.7 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
| 27 | c1ofgF_ |
|
not modelled |
96.7 |
16 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
| 28 | c1h6dL_ |
|
not modelled |
96.7 |
16 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
| 29 | c3nt5B_ |
|
not modelled |
96.7 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;
PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
|
| 30 | c3f4lF_ |
|
not modelled |
96.6 |
16 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative oxidoreductase yhhx;
PDBTitle: crystal structure of a probable oxidoreductase yhhx in2 triclinic form. northeast structural genomics target er647
|
| 31 | c3rbvA_ |
|
not modelled |
96.6 |
13 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar 3-ketoreductase;
PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
|
| 32 | d1zh8a1 |
|
not modelled |
96.6 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 33 | c3e9mC_ |
|
not modelled |
96.5 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from enterococcus2 faecalis
|
| 34 | c2o48X_ |
|
not modelled |
96.3 |
13 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
| 35 | c3dtyA_ |
|
not modelled |
96.2 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of an oxidoreductase from pseudomonas2 syringae
|
| 36 | d1ryda1 |
|
not modelled |
96.2 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 37 | c2ho3D_ |
|
not modelled |
96.2 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase, gfo/idh/moca family from2 streptococcus pneumoniae
|
| 38 | c3ceaA_ |
|
not modelled |
96.2 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
| 39 | c2q4eB_ |
|
not modelled |
95.8 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable oxidoreductase at4g09670;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at4g09670
|
| 40 | c3moiA_ |
|
not modelled |
95.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of the putative dehydrogenase from bordetella2 bronchiseptica rb50
|
| 41 | d1ydwa1 |
|
not modelled |
95.1 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 42 | c3oa0B_ |
|
not modelled |
94.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lipopolysaccaride biosynthesis protein wbpb;
PDBTitle: crystal structure of the wlba (wbpb) dehydrogenase from thermus2 thermophilus in complex with nad and udp-glcnaca
|
| 43 | c3oqbF_ |
|
not modelled |
94.8 |
20 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from bradyrhizobium2 japonicum usda 110
|
| 44 | c3fhlC_ |
|
not modelled |
94.6 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from bacteroides2 fragilis nctc 9343
|
| 45 | c1lc3A_ |
|
not modelled |
94.4 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:biliverdin reductase a;
PDBTitle: crystal structure of a biliverdin reductase enzyme-cofactor2 complex
|
| 46 | c1xeaD_ |
|
not modelled |
94.0 |
10 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of a gfo/idh/moca family oxidoreductase2 from vibrio cholerae
|
| 47 | c3q2kB_ |
|
not modelled |
94.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
|
| 48 | c1tltB_ |
|
not modelled |
93.6 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase (virulence factor mvim homolog);
PDBTitle: crystal structure of a putative oxidoreductase (virulence factor mvim2 homolog)
|
| 49 | c3oa2B_ |
|
not modelled |
93.6 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:wbpb;
PDBTitle: crystal structure of the wlba (wbpb) dehydrogenase from pseudomonas2 aeruginosa in complex with nad at 1.5 angstrom resolution
|
| 50 | c3ip3D_ |
|
not modelled |
93.6 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, putative;
PDBTitle: structure of putative oxidoreductase (tm_0425) from2 thermotoga maritima
|
| 51 | d1tlta1 |
|
not modelled |
93.4 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 52 | d1xeaa1 |
|
not modelled |
93.4 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 53 | c2ixaA_ |
|
not modelled |
93.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-n-acetylgalactosaminidase;
PDBTitle: a-zyme, n-acetylgalactosaminidase
|
| 54 | d1lc0a1 |
|
not modelled |
92.4 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 55 | c3evnA_ |
|
not modelled |
91.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of putative oxidoreductase from streptococcus2 agalactiae 2603v/r
|
| 56 | d1oi7a1 |
|
not modelled |
90.2 |
6 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 57 | c3v5nA_ |
|
not modelled |
89.7 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: the crystal structure of oxidoreductase from sinorhizobium meliloti
|
| 58 | c2zcuA_ |
|
not modelled |
89.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized oxidoreductase ytfg;
PDBTitle: crystal structure of a new type of nadph-dependent quinone2 oxidoreductase (qor2) from escherichia coli
|
| 59 | d1xgka_ |
|
not modelled |
86.9 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 60 | c2vrcD_ |
|
not modelled |
86.3 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:triphenylmethane reductase;
PDBTitle: crystal structure of the citrobacter sp. triphenylmethane2 reductase complexed with nadp(h)
|
| 61 | c2c20D_ |
|
not modelled |
81.5 |
24 |
PDB header:isomerase
Chain: D: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of udp-glucose 4-epimerase
|
| 62 | d2nu7a1 |
|
not modelled |
79.5 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 63 | c2v6gA_ |
|
not modelled |
79.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:progesterone 5-beta-reductase;
PDBTitle: structure of progesterone 5beta-reductase from digitalis2 lanata in complex with nadp
|
| 64 | d1wvga1 |
|
not modelled |
77.0 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 65 | c2gn9B_ |
|
not modelled |
74.5 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:udp-glcnac c6 dehydratase;
PDBTitle: crystal structure of udp-glcnac inverting 4,6-dehydratase in complex2 with nadp and udp-glc
|
| 66 | c1t2aC_ |
|
not modelled |
73.5 |
17 |
PDB header:structural genomics,lyase
Chain: C: PDB Molecule:gdp-mannose 4,6 dehydratase;
PDBTitle: crystal structure of human gdp-d-mannose 4,6-dehydratase
|
| 67 | d1t2aa_ |
|
not modelled |
73.5 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 68 | c2z1mC_ |
|
not modelled |
68.0 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:gdp-d-mannose dehydratase;
PDBTitle: crystal structure of gdp-d-mannose dehydratase from aquifex aeolicus2 vf5
|
| 69 | d1db3a_ |
|
not modelled |
67.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 70 | d1x1na1 |
|
not modelled |
65.2 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 71 | d1z45a2 |
|
not modelled |
64.6 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 72 | d1tz7a1 |
|
not modelled |
63.5 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 73 | d1eswa_ |
|
not modelled |
61.0 |
30 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 74 | d1rkxa_ |
|
not modelled |
60.7 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 75 | d1udca_ |
|
not modelled |
59.9 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 76 | c2qx7A_ |
|
not modelled |
57.5 |
16 |
PDB header:plant protein
Chain: A: PDB Molecule:eugenol synthase 1;
PDBTitle: structure of eugenol synthase from ocimum basilicum
|
| 77 | d2q46a1 |
|
not modelled |
55.9 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 78 | c2iodD_ |
|
not modelled |
55.4 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydroflavonol 4-reductase;
PDBTitle: binding of two substrate analogue molecules to2 dihydroflavonol-4-reductase alters the functional geometry3 of the catalytic site
|
| 79 | c3enkB_ |
|
not modelled |
54.8 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: 1.9a crystal structure of udp-glucose 4-epimerase from2 burkholderia pseudomallei
|
| 80 | c2p5uC_ |
|
not modelled |
53.7 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-2 epimerase complex with nad
|
| 81 | c3e48B_ |
|
not modelled |
53.2 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative nucleoside-diphosphate-sugar epimerase;
PDBTitle: crystal structure of a nucleoside-diphosphate-sugar epimerase2 (sav0421) from staphylococcus aureus, northeast structural genomics3 consortium target zr319
|
| 82 | c3mjsA_ |
|
not modelled |
52.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:amphb;
PDBTitle: structure of a-type ketoreductases from modular polyketide synthase
|
| 83 | c2pk3B_ |
|
not modelled |
52.2 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-6-deoxy-d-lyxo-4-hexulose reductase;
PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
|
| 84 | c3slgB_ |
|
not modelled |
51.6 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:pbgp3 protein;
PDBTitle: crystal structure of pbgp3 protein from burkholderia pseudomallei
|
| 85 | d1i24a_ |
|
not modelled |
51.5 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 86 | d2blla1 |
|
not modelled |
50.2 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 87 | d1n7ha_ |
|
not modelled |
48.2 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 88 | c2q1wC_ |
|
not modelled |
47.6 |
23 |
PDB header:sugar binding protein
Chain: C: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmh in2 complex with nad+
|
| 89 | d1euca1 |
|
not modelled |
47.3 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 90 | c3qvoA_ |
|
not modelled |
46.6 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nmra family protein;
PDBTitle: structure of a rossmann-fold nad(p)-binding family protein from2 shigella flexneri.
|
| 91 | c2p2sA_ |
|
not modelled |
44.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase (yp_050235.1) from2 erwinia carotovora atroseptica scri1043 at 1.25 a resolution
|
| 92 | c3r6dA_ |
|
not modelled |
43.9 |
17 |
PDB header:lyase, isomerase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of nad-dependent epimerase/dehydratase from2 veillonella parvula dsm 2008 with cz-methylated lysine
|
| 93 | d1bxka_ |
|
not modelled |
43.8 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 94 | d1orra_ |
|
not modelled |
42.9 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 95 | c1e5lA_ |
|
not modelled |
41.3 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine reductase;
PDBTitle: apo saccharopine reductase from magnaporthe grisea
|
| 96 | c3h2sA_ |
|
not modelled |
40.3 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative nadh-flavin reductase;
PDBTitle: crystal structure of the q03b84 protein from lactobacillus2 casei. northeast structural genomics consortium target3 lcr19.
|
| 97 | c3ay3C_ |
|
not modelled |
38.3 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of glucuronic acid dehydrogeanse from2 chromohalobacter salexigens
|
| 98 | d1gy8a_ |
|
not modelled |
35.1 |
9 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 99 | c2p4hX_ |
|
not modelled |
35.1 |
30 |
PDB header:plant protein
Chain: X: PDB Molecule:vestitone reductase;
PDBTitle: crystal structure of vestitone reductase from alfalfa2 (medicago sativa l.)
|
| 100 | c1j5pA_ |
|
not modelled |
34.8 |
6 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate dehydrogenase;
PDBTitle: crystal structure of aspartate dehydrogenase (tm1643) from thermotoga2 maritima at 1.9 a resolution
|
| 101 | c2axqA_ |
|
not modelled |
34.8 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
| 102 | d1vl0a_ |
|
not modelled |
34.2 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 103 | c3dqpA_ |
|
not modelled |
32.5 |
34 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase ylbe;
PDBTitle: crystal structure of the oxidoreductase ylbe from2 lactococcus lactis, northeast structural genomics3 consortium target kr121.
|
| 104 | c3dhnA_ |
|
not modelled |
32.4 |
33 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of the putative epimerase q89z24_bactn2 from bacteroides thetaiotaomicron. northeast structural3 genomics consortium target btr310.
|
| 105 | c2ggsB_ |
|
not modelled |
32.3 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:273aa long hypothetical dtdp-4-dehydrorhamnose
PDBTitle: crystal structure of hypothetical dtdp-4-dehydrorhamnose2 reductase from sulfolobus tokodaii
|
| 106 | c3ew7A_ |
|
not modelled |
31.8 |
28 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lmo0794 protein;
PDBTitle: crystal structure of the lmo0794 protein from listeria2 monocytogenes. northeast structural genomics consortium3 target lmr162.
|
| 107 | c1z7eC_ |
|
not modelled |
31.3 |
11 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein arna;
PDBTitle: crystal structure of full length arna
|
| 108 | c2pzlB_ |
|
not modelled |
30.8 |
16 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme2 wbmg in complex with nad and udp
|
| 109 | c3th6B_ |
|
not modelled |
30.5 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from rhipicephalus2 (boophilus) microplus.
|
| 110 | d1n2sa_ |
|
not modelled |
30.3 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 111 | d1ebfa1 |
|
not modelled |
29.7 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 112 | d1rpna_ |
|
not modelled |
29.5 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 113 | c3rfxB_ |
|
not modelled |
29.2 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uronate dehydrogenase;
PDBTitle: crystal structure of uronate dehydrogenase from agrobacterium2 tumefaciens, y136a mutant complexed with nad
|
| 114 | c3sc6F_ |
|
not modelled |
27.8 |
30 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: 2.65 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 reductase (rfbd) from bacillus anthracis str. ames in complex with3 nadp
|
| 115 | d1nnxa_ |
|
not modelled |
27.5 |
60 |
Fold:OB-fold
Superfamily:Hypothetical protein YgiW
Family:Hypothetical protein YgiW |
| 116 | c2vz8B_ |
|
not modelled |
27.4 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase;
PDBTitle: crystal structure of mammalian fatty acid synthase
|
| 117 | c1n7gB_ |
|
not modelled |
27.1 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:gdp-d-mannose-4,6-dehydratase;
PDBTitle: crystal structure of the gdp-mannose 4,6-dehydratase2 ternary complex with nadph and gdp-rhamnose.
|
| 118 | d1ek6a_ |
|
not modelled |
27.1 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 119 | c2ysrA_ |
|
not modelled |
26.6 |
24 |
PDB header:signaling protein
Chain: A: PDB Molecule:dep domain-containing protein 1;
PDBTitle: solution structure of the dep domain from human dep domain-2 containing protein 1
|
| 120 | c3kxqB_ |
|
not modelled |
26.2 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from bartonella2 henselae at 1.6a resolution
|