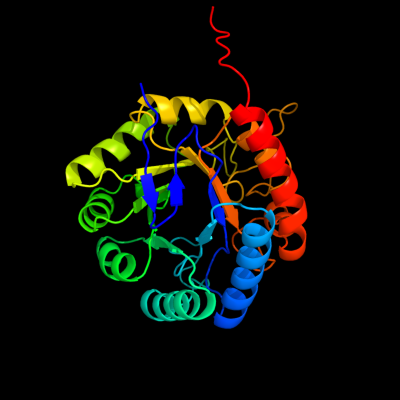
| 1 | d1o60a_
|
|
 |
100.0 |
78 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
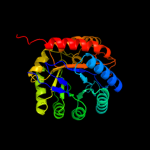
| 2 | d1d9ea_
|
|
 |
100.0 |
99 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
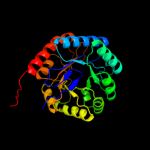
| 3 | c3stgA_
|
|
 |
100.0 |
70 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
|
| 4 | c3sz8D_
|
|
 |
100.0 |
66 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 2;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
|
| 5 | d2a21a1
|
|
 |
100.0 |
46 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 6 | c1xuzA_
|
|
 |
100.0 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:polysialic acid capsule biosynthesis protein siac;
PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
|
| 7 | c1vliA_
|
|
 |
100.0 |
12 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:spore coat polysaccharide biosynthesis protein spse;
PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
|
| 8 | d1vlia2
|
|
 |
100.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 9 | d2zdra2
|
|
 |
100.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:NeuB-like |
| 10 | c3fs2A_
|
|
 |
100.0 |
40 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate2 aldolase from bruciella melitensis at 1.85a resolution
|
| 11 | d1vr6a1
|
|
 |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 12 | c1zcoA_
|
|
 |
100.0 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphoheptonate aldolase;
PDBTitle: crystal structure of pyrococcus furiosus 3-deoxy-d-arabino-2 heptulosonate 7-phosphate synthase
|
| 13 | c1vs1B_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:3-deoxy-7-phosphoheptulonate synthase;
PDBTitle: crystal structure of 3-deoxy-d-arabino-heptulosonate-7-2 phosphate synthase (dahp synthase) from aeropyrum pernix3 in complex with mn2+ and pep
|
| 14 | c3pg8B_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: truncated form of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase2 from thermotoga maritima
|
| 15 | c3t4cD_
|
|
 |
100.0 |
42 |
PDB header:transferase
Chain: D: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase 1;
PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia ambifaria
|
| 16 | c3nvtA_
|
|
 |
100.0 |
26 |
PDB header:transferase/isomerase
Chain: A: PDB Molecule:3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;
PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
|
| 17 | c3g8rA_
|
|
 |
100.0 |
9 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:probable spore coat polysaccharide biosynthesis protein e;
PDBTitle: crystal structure of putative spore coat polysaccharide biosynthesis2 protein e from chromobacterium violaceum atcc 12472
|
| 18 | c1ofaB_
|
|
 |
100.0 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: crystal structure of the tyrosine-regulated2 3-deoxy-d-arabino-heptulosonate-7-phosphate synthase3 from saccharomyces cerevisiae in complex with4 phosphoenolpyruvate and cobalt(ii)
|
| 19 | d1of8a_
|
|
 |
100.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 20 | d1n8fa_
|
|
 |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I DAHP synthetase |
| 21 | c3tqkA_ |
|
not modelled |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: structure of phospho-2-dehydro-3-deoxyheptonate aldolase from2 francisella tularensis schu s4
|
| 22 | c2y5sA_ |
|
not modelled |
98.3 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
| 23 | c1tx2A_ |
|
not modelled |
98.1 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 24 | d1tx2a_ |
|
not modelled |
98.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 25 | d1rd5a_ |
|
not modelled |
97.9 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 26 | c2yciX_ |
|
not modelled |
97.9 |
11 |
PDB header:transferase
Chain: X: PDB Molecule:5-methyltetrahydrofolate corrinoid/iron sulfur protein
PDBTitle: methyltransferase native
|
| 27 | d1f6ya_ |
|
not modelled |
97.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 28 | d1wbha1 |
|
not modelled |
97.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 29 | d1ajza_ |
|
not modelled |
97.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 30 | c2vp8A_ |
|
not modelled |
97.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase 2;
PDBTitle: structure of mycobacterium tuberculosis rv1207
|
| 31 | d1wa3a1 |
|
not modelled |
97.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 32 | d1vhca_ |
|
not modelled |
97.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 33 | c1piiA_ |
|
not modelled |
97.4 |
10 |
PDB header:bifunctional(isomerase and synthase)
Chain: A: PDB Molecule:n-(5'phosphoribosyl)anthranilate isomerase;
PDBTitle: three-dimensional structure of the bifunctional enzyme2 phosphoribosylanthranilate isomerase:3 indoleglycerolphosphate synthase from escherichia coli4 refined at 2.0 angstroms resolution
|
| 34 | c3noyA_ |
|
not modelled |
97.3 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
| 35 | d1ad1a_ |
|
not modelled |
97.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 36 | c1zlpA_ |
|
not modelled |
97.3 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
| 37 | c3gndC_ |
|
not modelled |
97.2 |
8 |
PDB header:lyase
Chain: C: PDB Molecule:aldolase lsrf;
PDBTitle: crystal structure of e. coli lsrf in complex with ribulose-5-phosphate
|
| 38 | c2h9aB_ |
|
not modelled |
97.2 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase, iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 39 | c3ih1A_ |
|
not modelled |
97.2 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 40 | d1d3ga_ |
|
not modelled |
97.1 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 41 | d1to3a_ |
|
not modelled |
97.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 42 | d1a53a_ |
|
not modelled |
97.0 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 43 | c2bmbA_ |
|
not modelled |
97.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase3 from saccharomyces cerevisiae
|
| 44 | d3bofa1 |
|
not modelled |
97.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 45 | c3jrkG_ |
|
not modelled |
97.0 |
13 |
PDB header:lyase
Chain: G: PDB Molecule:tagatose 1,6-diphosphate aldolase 2;
PDBTitle: a putative tagatose 1,6-diphosphate aldolase from streptococcus2 pyogenes
|
| 46 | c3tr9A_ |
|
not modelled |
96.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 47 | d1vc4a_ |
|
not modelled |
96.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 48 | d1uuma_ |
|
not modelled |
96.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 49 | c3eooL_ |
|
not modelled |
96.9 |
15 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 50 | d1eyea_ |
|
not modelled |
96.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 51 | d2fyma1 |
|
not modelled |
96.7 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 52 | c3oqbF_ |
|
not modelled |
96.7 |
13 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from bradyrhizobium2 japonicum usda 110
|
| 53 | d1s2wa_ |
|
not modelled |
96.6 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 54 | c3qjaA_ |
|
not modelled |
96.6 |
8 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of the mycobacterium tuberculosis indole-3-glycerol2 phosphate synthase (trpc) in apo form
|
| 55 | d1mxsa_ |
|
not modelled |
96.6 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 56 | d1wv2a_ |
|
not modelled |
96.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
| 57 | c2y0fD_ |
|
not modelled |
96.5 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
| 58 | c2dzaA_ |
|
not modelled |
96.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
| 59 | d1piia2 |
|
not modelled |
96.5 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 60 | c2c3zA_ |
|
not modelled |
96.4 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
| 61 | c3tqpA_ |
|
not modelled |
96.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:enolase;
PDBTitle: structure of an enolase (eno) from coxiella burnetii
|
| 62 | c2ekcA_ |
|
not modelled |
96.4 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
| 63 | d1muma_ |
|
not modelled |
96.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 64 | c2vefB_ |
|
not modelled |
96.4 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:dihydropteroate synthase;
PDBTitle: dihydropteroate synthase from streptococcus pneumoniae
|
| 65 | c3v5nA_ |
|
not modelled |
96.4 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: the crystal structure of oxidoreductase from sinorhizobium meliloti
|
| 66 | c3eb2A_ |
|
not modelled |
96.4 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:putative dihydrodipicolinate synthetase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 2.0a resolution
|
| 67 | d1qopa_ |
|
not modelled |
96.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 68 | c2v82A_ |
|
not modelled |
96.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
| 69 | d2akza1 |
|
not modelled |
96.2 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 70 | c3moiA_ |
|
not modelled |
96.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable dehydrogenase;
PDBTitle: the crystal structure of the putative dehydrogenase from bordetella2 bronchiseptica rb50
|
| 71 | c3k13A_ |
|
not modelled |
96.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
|
| 72 | c2r8wB_ |
|
not modelled |
96.1 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
| 73 | d1ka9f_ |
|
not modelled |
96.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 74 | d2b7oa1 |
|
not modelled |
96.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class-II DAHP synthetase |
| 75 | d1p0ka_ |
|
not modelled |
95.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 76 | c2ftpA_ |
|
not modelled |
95.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
| 77 | c1l8pC_ |
|
not modelled |
95.9 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:enolase 1;
PDBTitle: mg-phosphonoacetohydroxamate complex of s39a yeast enolase 1
|
| 78 | c3ezyB_ |
|
not modelled |
95.9 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of probable dehydrogenase tm_0414 from2 thermotoga maritima
|
| 79 | c3db2C_ |
|
not modelled |
95.8 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
| 80 | c3rbvA_ |
|
not modelled |
95.7 |
13 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar 3-ketoreductase;
PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
|
| 81 | d1pdza1 |
|
not modelled |
95.6 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 82 | d1i4na_ |
|
not modelled |
95.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 83 | c3qn3B_ |
|
not modelled |
95.6 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:enolase;
PDBTitle: phosphopyruvate hydratase from campylobacter jejuni.
|
| 84 | c2akmA_ |
|
not modelled |
95.6 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:gamma enolase;
PDBTitle: fluoride inhibition of enolase: crystal structure of the2 inhibitory complex
|
| 85 | c2v9dB_ |
|
not modelled |
95.6 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to2 the dihydrodipicolinic acid synthase family from e. coli3 k12
|
| 86 | d2ptza1 |
|
not modelled |
95.5 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 87 | c3qc3B_ |
|
not modelled |
95.4 |
10 |
PDB header:isomerase
Chain: B: PDB Molecule:d-ribulose-5-phosphate-3-epimerase;
PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
|
| 88 | c2nvwB_ |
|
not modelled |
95.4 |
14 |
PDB header:transcription
Chain: B: PDB Molecule:galactose/lactose metabolism regulatory protein
PDBTitle: crystal sctucture of transcriptional regulator gal80p from2 kluyveromymes lactis
|
| 89 | c3ceaA_ |
|
not modelled |
95.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
| 90 | c3euwB_ |
|
not modelled |
95.4 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:myo-inositol dehydrogenase;
PDBTitle: crystal structure of a myo-inositol dehydrogenase from corynebacterium2 glutamicum atcc 13032
|
| 91 | c2rfgB_ |
|
not modelled |
95.3 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from hahella2 chejuensis at 1.5a resolution
|
| 92 | c3q2kB_ |
|
not modelled |
95.3 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of the wlba dehydrogenase from bordetella pertussis2 in complex with nadh and udp-glcnaca
|
| 93 | c3fluD_ |
|
not modelled |
95.3 |
14 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from the pathogen2 neisseria meningitidis
|
| 94 | c3uj2C_ |
|
not modelled |
95.3 |
16 |
PDB header:lyase
Chain: C: PDB Molecule:enolase 1;
PDBTitle: crystal structure of an enolase from anaerostipes caccae (efi target2 efi-502054) with bound mg and sulfate
|
| 95 | d1xkya1 |
|
not modelled |
95.3 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 96 | c3q58A_ |
|
not modelled |
95.2 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
|
| 97 | d1j5ta_ |
|
not modelled |
95.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 98 | c2q4eB_ |
|
not modelled |
95.2 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable oxidoreductase at4g09670;
PDBTitle: ensemble refinement of the protein crystal structure of gene product2 from arabidopsis thaliana at4g09670
|
| 99 | d1f76a_ |
|
not modelled |
95.2 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 100 | d1thfd_ |
|
not modelled |
95.2 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
| 101 | c1xeaD_ |
|
not modelled |
95.1 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of a gfo/idh/moca family oxidoreductase2 from vibrio cholerae
|
| 102 | c3lyeA_ |
|
not modelled |
95.1 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 103 | c3noeA_ |
|
not modelled |
95.1 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from pseudomonas2 aeruginosa
|
| 104 | d1o5ka_ |
|
not modelled |
95.1 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 105 | c3l2iB_ |
|
not modelled |
95.0 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
|
| 106 | d1w6ta1 |
|
not modelled |
95.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:Enolase |
| 107 | c3qtpB_ |
|
not modelled |
95.0 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:enolase 1;
PDBTitle: crystal structure analysis of entamoeba histolytica enolase
|
| 108 | c3pueA_ |
|
not modelled |
95.0 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
| 109 | c3bi8A_ |
|
not modelled |
95.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: structure of dihydrodipicolinate synthase from clostridium2 botulinum
|
| 110 | c1h6dL_ |
|
not modelled |
95.0 |
15 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
| 111 | d1h1ya_ |
|
not modelled |
94.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:D-ribulose-5-phosphate 3-epimerase |
| 112 | c1ofgF_ |
|
not modelled |
94.9 |
15 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
| 113 | c2o48X_ |
|
not modelled |
94.9 |
13 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:dimeric dihydrodiol dehydrogenase;
PDBTitle: crystal structure of mammalian dimeric dihydrodiol dehydrogenase
|
| 114 | c1zfjA_ |
|
not modelled |
94.8 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
| 115 | d1xi3a_ |
|
not modelled |
94.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Thiamin phosphate synthase
Family:Thiamin phosphate synthase |
| 116 | c1zh8B_ |
|
not modelled |
94.8 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
| 117 | c2fptA_ |
|
not modelled |
94.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydroorotate dehydrogenase, mitochondrial;
PDBTitle: dual binding mode of a novel series of dhodh inhibitors
|
| 118 | c3otrC_ |
|
not modelled |
94.8 |
11 |
PDB header:lyase
Chain: C: PDB Molecule:enolase;
PDBTitle: 2.75 angstrom crystal structure of enolase 1 from toxoplasma gondii
|
| 119 | c3igsB_ |
|
not modelled |
94.8 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase 2;
PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
|
| 120 | c2qjhH_ |
|
not modelled |
94.8 |
16 |
PDB header:lyase
Chain: H: PDB Molecule:putative aldolase mj0400;
PDBTitle: m. jannaschii adh synthase covalently bound to2 dihydroxyacetone phosphate
|