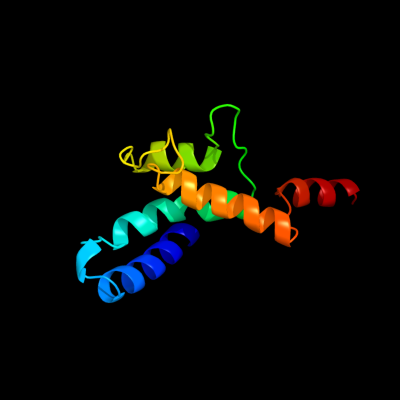
1 c2ht2B_
44.4
17
PDB header: membrane proteinChain: B: PDB Molecule: h(+)/cl(-) exchange transporter clca;PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
2 d1otsa_
41.4
16
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel3 c2yvxD_
28.6
17
PDB header: transport proteinChain: D: PDB Molecule: mg2+ transporter mgte;PDBTitle: crystal structure of magnesium transporter mgte
4 d1kpla_
14.7
16
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel5 d1fx8a_
12.2
13
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like6 c1ldaA_
12.2
13
PDB header: transport proteinChain: A: PDB Molecule: glycerol uptake facilitator protein;PDBTitle: crystal structure of the e. coli glycerol facilitator (glpf) without2 substrate glycerol
7 c2yggA_
11.7
9
PDB header: metal binding protein/transport proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: complex of cambr and cam
8 c1ymgA_
11.7
8
PDB header: membrane proteinChain: A: PDB Molecule: lens fiber major intrinsic protein;PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
9 d1ymga1
11.7
8
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like10 d1ciya3
11.4
10
Fold: Toxins' membrane translocation domainsSuperfamily: delta-Endotoxin (insectocide), N-terminal domainFamily: delta-Endotoxin (insectocide), N-terminal domain11 d2nefa_
11.4
23
Fold: Regulatory factor NefSuperfamily: Regulatory factor NefFamily: Regulatory factor Nef12 c3nd0A_
11.2
13
PDB header: transport proteinChain: A: PDB Molecule: sll0855 protein;PDBTitle: x-ray crystal structure of a slow cyanobacterial cl-/h+ antiporter
13 c2kncB_
11.2
17
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
14 c3rbbA_
10.7
20
PDB header: viral protein, protein bindingChain: A: PDB Molecule: protein nef;PDBTitle: hiv-1 nef protein in complex with engineered hck sh3 domain
15 d1e12a_
10.5
11
Fold: Family A G protein-coupled receptor-likeSuperfamily: Family A G protein-coupled receptor-likeFamily: Bacteriorhodopsin-like16 d1iwga8
10.2
13
Fold: Multidrug efflux transporter AcrB transmembrane domainSuperfamily: Multidrug efflux transporter AcrB transmembrane domainFamily: Multidrug efflux transporter AcrB transmembrane domain17 d1rc2a_
10.1
12
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like18 d1j4na_
9.9
10
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like19 d2r6gf1
9.6
10
Fold: MalF N-terminal region-likeSuperfamily: MalF N-terminal region-likeFamily: MalF N-terminal region-like20 c2rmzA_
9.4
17
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-3;PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
21 c3pltB_
not modelled
9.2
14
PDB header: structural proteinChain: B: PDB Molecule: sphingolipid long chain base-responsive protein lsp1;PDBTitle: crystal structure of lsp1 from saccharomyces cerevisiae
22 c3k3gA_
not modelled
8.8
14
PDB header: transport proteinChain: A: PDB Molecule: urea transporter;PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
23 c2jagA_
not modelled
8.6
11
PDB header: membrane proteinChain: A: PDB Molecule: halorhodopsin;PDBTitle: l1-intermediate of halorhodopsin t203v
24 d1pw4a_
not modelled
8.4
7
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter25 c3hfwA_
not modelled
8.3
13
PDB header: hydrolaseChain: A: PDB Molecule: protein adp-ribosylarginine hydrolase;PDBTitle: crystal structure of human adp-ribosylhydrolase 1 (harh1)
26 c2wocA_
not modelled
8.3
25
PDB header: hydrolaseChain: A: PDB Molecule: adp-ribosyl-[dinitrogen reductase] glycohydrolase;PDBTitle: crystal structure of the dinitrogenase reductase-activating2 glycohydrolase (drag) from rhodospirillum rubrum
27 c3msqC_
not modelled
8.2
10
PDB header: biosynthetic proteinChain: C: PDB Molecule: putative ubiquinone biosynthesis protein;PDBTitle: crystal structure of a putative ubiquinone biosynthesis protein2 (npun02000094) from nostoc punctiforme pcc 73102 at 2.85 a resolution
28 c2kbvA_
not modelled
8.1
21
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: structural and functional analysis of tm xi of the nhe12 isoform of the na+/h+ exchanger
29 c1i5pA_
not modelled
7.8
14
PDB header: toxinChain: A: PDB Molecule: pesticidial crystal protein cry2aa;PDBTitle: insecticidal crystal protein cry2aa
30 c1iijA_
not modelled
7.7
21
PDB header: signaling proteinChain: A: PDB Molecule: erbb-2 receptor protein-tyrosine kinase;PDBTitle: solution structure of the neu/erbb-2 membrane spanning2 segment
31 c3k07A_
not modelled
7.6
10
PDB header: transport proteinChain: A: PDB Molecule: cation efflux system protein cusa;PDBTitle: crystal structure of cusa
32 c3g9dB_
not modelled
7.6
25
PDB header: hydrolaseChain: B: PDB Molecule: dinitrogenase reductase activactingPDBTitle: crystal structure glycohydrolase
33 c2kdcC_
not modelled
7.5
5
PDB header: transferaseChain: C: PDB Molecule: diacylglycerol kinase;PDBTitle: nmr solution structure of e. coli diacylglycerol kinase2 (dagk) in dpc micelles
34 c1iojA_
not modelled
7.4
21
PDB header: apolipoproteinChain: A: PDB Molecule: apoc-i;PDBTitle: human apolipoprotein c-i, nmr, 18 structures
35 d1i5pa3
not modelled
7.3
13
Fold: Toxins' membrane translocation domainsSuperfamily: delta-Endotoxin (insectocide), N-terminal domainFamily: delta-Endotoxin (insectocide), N-terminal domain36 c1ciyA_
not modelled
7.0
9
PDB header: toxinChain: A: PDB Molecule: cryia(a);PDBTitle: insecticidal toxin: structure and channel formation
37 c2b5fD_
not modelled
7.0
12
PDB header: transport protein,membrane proteinChain: D: PDB Molecule: aquaporin;PDBTitle: crystal structure of the spinach aquaporin sopip2;1 in an2 open conformation to 3.9 resolution
38 c2kncA_
not modelled
6.7
17
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
39 d2pw6a1
not modelled
6.6
7
Fold: Phosphorylase/hydrolase-likeSuperfamily: LigB-likeFamily: LigB-like40 c2qtyB_
not modelled
6.6
17
PDB header: hydrolaseChain: B: PDB Molecule: poly(adp-ribose) glycohydrolase arh3;PDBTitle: crystal structure of mouse adp-ribosylhydrolase 3 (marh3)
41 c2yzwA_
not modelled
6.5
20
PDB header: hydrolaseChain: A: PDB Molecule: adp-ribosylglycohydrolase;PDBTitle: adp-ribosylglycohydrolase-related protein complex
42 c3ik5A_
not modelled
6.4
21
PDB header: viral protein/signaling proteinChain: A: PDB Molecule: protein nef;PDBTitle: sivmac239 nef in complex with tcr zeta itam 1 polypeptide2 (a63-r80)
43 c2k1lB_
not modelled
6.3
28
PDB header: signaling proteinChain: B: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
44 c2k1kA_
not modelled
6.3
28
PDB header: signaling proteinChain: A: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
45 c2k1kB_
not modelled
6.3
28
PDB header: signaling proteinChain: B: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
46 c2k1lA_
not modelled
6.3
28
PDB header: signaling proteinChain: A: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
47 d1t5ja_
not modelled
6.3
21
Fold: ADP-ribosylglycohydrolaseSuperfamily: ADP-ribosylglycohydrolaseFamily: ADP-ribosylglycohydrolase48 c3t6gB_
not modelled
6.2
7
PDB header: signaling protein, cell adhesionChain: B: PDB Molecule: breast cancer anti-estrogen resistance protein 1;PDBTitle: structure of the complex between nsp3 (shep1) and p130cas
49 c2c9kA_
not modelled
6.0
13
PDB header: toxinChain: A: PDB Molecule: pesticidal crystal protein cry4aa;PDBTitle: structure of the functional form of the mosquito-larvicidal2 cry4aa toxin from bacillus thuringiensis at 2.8 a3 resolution
50 d1tuka1
not modelled
5.3
25
Fold: Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albuminSuperfamily: Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albuminFamily: Plant lipid-transfer and hydrophobic proteins51 d2bida_
not modelled
5.3
10
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death52 d1t01a1
not modelled
5.2
12
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin53 c2y69Q_
not modelled
5.2
6
PDB header: electron transportChain: Q: PDB Molecule: cytochrome c oxidase subunit 4 isoform 1;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
54 d1xw8a_
not modelled
5.1
15
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: LamB/YcsF-like