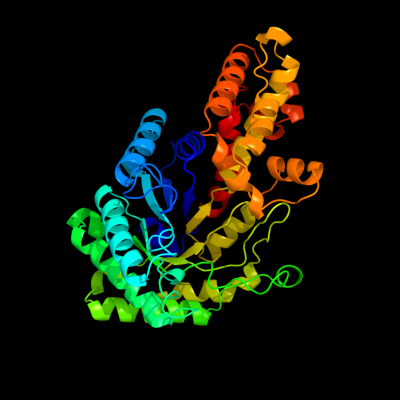
| 1 | d2fiqa1
|
|
 |
100.0 |
96 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:GatZ-like |
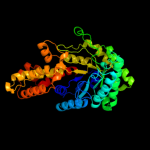
| 2 | c3q94B_
|
|
 |
99.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase, class ii;
PDBTitle: the crystal structure of fructose 1,6-bisphosphate aldolase from2 bacillus anthracis str. 'ames ancestor'
|
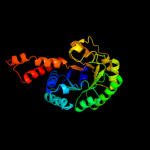
| 3 | d1rvga_
|
|
 |
99.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 4 | d1dosa_
|
|
 |
99.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 5 | d1gvfa_
|
|
 |
99.9 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 6 | c3c52B_
|
|
 |
99.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: class ii fructose-1,6-bisphosphate aldolase from2 helicobacter pylori in complex with3 phosphoglycolohydroxamic acid, a competitive inhibitor
|
| 7 | c3elfA_
|
|
 |
99.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: structural characterization of tetrameric mycobacterium tuberculosis2 fructose 1,6-bisphosphate aldolase - substrate binding and catalysis3 mechanism of a class iia bacterial aldolase
|
| 8 | c3qm3C_
|
|
 |
99.9 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:fructose-bisphosphate aldolase;
PDBTitle: 1.85 angstrom resolution crystal structure of fructose-bisphosphate2 aldolase (fba) from campylobacter jejuni
|
| 9 | c2iswB_
|
|
 |
99.9 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-1,6-bisphosphate aldolase;
PDBTitle: structure of giardia fructose-1,6-biphosphate aldolase in2 complex with phosphoglycolohydroxamate
|
| 10 | c3pm6B_
|
|
 |
99.9 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-bisphosphate aldolase;
PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
|
| 11 | d1f6ya_
|
|
 |
95.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 12 | c3bolB_
|
|
 |
95.8 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
| 13 | d3bofa1
|
|
 |
95.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
| 14 | c1rr2A_
|
|
 |
94.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
| 15 | d1muma_
|
|
 |
93.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 16 | c2h9aA_
|
|
 |
93.1 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carbon monoxide dehydrogenase corrinoid/iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 17 | c2yciX_
|
|
 |
91.9 |
20 |
PDB header:transferase
Chain: X: PDB Molecule:5-methyltetrahydrofolate corrinoid/iron sulfur protein
PDBTitle: methyltransferase native
|
| 18 | c3k13A_
|
|
 |
91.4 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:5-methyltetrahydrofolate-homocysteine methyltransferase;
PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
|
| 19 | c2dzaA_
|
|
 |
91.2 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of dihydropteroate synthase from thermus2 thermophilus hb8 in complex with 4-aminobenzoate
|
| 20 | c2nx9B_
|
|
 |
90.8 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:oxaloacetate decarboxylase 2, subunit alpha;
PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
|
| 21 | c2vp8A_ |
|
not modelled |
90.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase 2;
PDBTitle: structure of mycobacterium tuberculosis rv1207
|
| 22 | c2ze3A_ |
|
not modelled |
89.5 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 23 | c2qiwA_ |
|
not modelled |
88.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
| 24 | c1ps9A_ |
|
not modelled |
86.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-2 dienoyl coa reductase
|
| 25 | d1tx2a_ |
|
not modelled |
84.5 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 26 | c1tx2A_ |
|
not modelled |
84.5 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:dhps, dihydropteroate synthase;
PDBTitle: dihydropteroate synthetase, with bound inhibitor manic, from bacillus2 anthracis
|
| 27 | c3hf3A_ |
|
not modelled |
83.2 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chromate reductase;
PDBTitle: old yellow enzyme from thermus scotoductus sa-01
|
| 28 | c3b8iF_ |
|
not modelled |
83.0 |
18 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
| 29 | c3cqkB_ |
|
not modelled |
83.0 |
9 |
PDB header:isomerase
Chain: B: PDB Molecule:l-ribulose-5-phosphate 3-epimerase ulae;
PDBTitle: crystal structure of l-xylulose-5-phosphate 3-epimerase ulae (form b)2 complex with zn2+ and sulfate
|
| 30 | d1eyea_ |
|
not modelled |
82.4 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 31 | d1vhna_ |
|
not modelled |
80.8 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 32 | d1ps9a1 |
|
not modelled |
79.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 33 | d1ad1a_ |
|
not modelled |
77.9 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 34 | c3lhlA_ |
|
not modelled |
77.2 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative agmatinase;
PDBTitle: crystal structure of a putative agmatinase from clostridium difficile
|
| 35 | d1z41a1 |
|
not modelled |
75.2 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 36 | c3k2gA_ |
|
not modelled |
75.1 |
13 |
PDB header:resiniferatoxin binding protein
Chain: A: PDB Molecule:resiniferatoxin-binding, phosphotriesterase-
PDBTitle: crystal structure of a resiniferatoxin-binding protein from2 rhodobacter sphaeroides
|
| 37 | d1vyra_ |
|
not modelled |
72.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 38 | d1ajza_ |
|
not modelled |
71.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
| 39 | c3d0kA_ |
|
not modelled |
71.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative poly(3-hydroxybutyrate) depolymerase lpqc;
PDBTitle: crystal structure of the lpqc, poly(3-hydroxybutyrate) depolymerase2 from bordetella parapertussis
|
| 40 | c1vraB_ |
|
not modelled |
69.9 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:arginine biosynthesis bifunctional protein argj;
PDBTitle: crystal structure of arginine biosynthesis bifunctional protein argj2 (10175521) from bacillus halodurans at 2.00 a resolution
|
| 41 | c2y5sA_ |
|
not modelled |
68.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: crystal structure of burkholderia cenocepacia dihydropteroate2 synthase complexed with 7,8-dihydropteroate.
|
| 42 | c3inpA_ |
|
not modelled |
68.2 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:d-ribulose-phosphate 3-epimerase;
PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
|
| 43 | d1gq6a_ |
|
not modelled |
67.8 |
19 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 44 | c3it4B_ |
|
not modelled |
67.2 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:arginine biosynthesis bifunctional protein argj
PDBTitle: the crystal structure of ornithine acetyltransferase from2 mycobacterium tuberculosis (rv1653) at 1.7 a
|
| 45 | c3l5aA_ |
|
not modelled |
67.2 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh/flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of a probable nadh-dependent flavin oxidoreductase2 from staphylococcus aureus
|
| 46 | c3nioF_ |
|
not modelled |
66.9 |
19 |
PDB header:hydrolase
Chain: F: PDB Molecule:guanidinobutyrase;
PDBTitle: crystal structure of pseudomonas aeruginosa guanidinobutyrase
|
| 47 | c2gq8A_ |
|
not modelled |
65.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, fmn-binding;
PDBTitle: structure of sye1, an oye homologue from s. ondeidensis, in complex2 with p-hydroxyacetophenone
|
| 48 | d1pq3a_ |
|
not modelled |
63.7 |
19 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 49 | d1xfka_ |
|
not modelled |
63.6 |
18 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 50 | d2a0ma1 |
|
not modelled |
62.6 |
26 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 51 | d2ceva_ |
|
not modelled |
61.4 |
18 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 52 | d2glka1 |
|
not modelled |
59.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 53 | c3atyA_ |
|
not modelled |
57.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:prostaglandin f2a synthase;
PDBTitle: crystal structure of tcoye
|
| 54 | d2ihta3 |
|
not modelled |
55.5 |
27 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
| 55 | c3r4iB_ |
|
not modelled |
55.2 |
6 |
PDB header:lyase
Chain: B: PDB Molecule:citrate lyase;
PDBTitle: crystal structure of a citrate lyase (bxe_b2899) from burkholderia2 xenovorans lb400 at 2.24 a resolution
|
| 56 | d1woha_ |
|
not modelled |
54.4 |
21 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 57 | c1cqxB_ |
|
not modelled |
50.3 |
25 |
PDB header:lipid binding protein
Chain: B: PDB Molecule:flavohemoprotein;
PDBTitle: crystal structure of the flavohemoglobin from alcaligenes eutrophus at2 1.75 a resolution
|
| 58 | d1ujqa_ |
|
not modelled |
49.5 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 59 | d1t4ba1 |
|
not modelled |
48.6 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 60 | d2csua1 |
|
not modelled |
47.9 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 61 | c3nipB_ |
|
not modelled |
47.9 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:3-guanidinopropionase;
PDBTitle: crystal structure of pseudomonas aeruginosa guanidinopropionase2 complexed with 1,6-diaminohexane
|
| 62 | d1qi9a_ |
|
not modelled |
47.3 |
17 |
Fold:Acid phosphatase/Vanadium-dependent haloperoxidase
Superfamily:Acid phosphatase/Vanadium-dependent haloperoxidase
Family:Haloperoxidase (bromoperoxidase) |
| 63 | d1gwja_ |
|
not modelled |
46.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 64 | d1m6ja_ |
|
not modelled |
46.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 65 | c3qxbB_ |
|
not modelled |
46.1 |
8 |
PDB header:isomerase
Chain: B: PDB Molecule:putative xylose isomerase;
PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
|
| 66 | d1yl7a1 |
|
not modelled |
45.7 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
| 67 | d1icpa_ |
|
not modelled |
45.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 68 | d1h6za1 |
|
not modelled |
43.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 69 | d1t9ba3 |
|
not modelled |
43.4 |
21 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase PP module |
| 70 | c2h9aB_ |
|
not modelled |
43.3 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase, iron-
PDBTitle: corrinoid iron-sulfur protein
|
| 71 | c3gr7A_ |
|
not modelled |
43.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal2 crystal form
|
| 72 | d1vz6a_ |
|
not modelled |
42.5 |
20 |
Fold:DmpA/ArgJ-like
Superfamily:DmpA/ArgJ-like
Family:ArgJ-like |
| 73 | d1oyaa_ |
|
not modelled |
41.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 74 | c2o1tB_ |
|
not modelled |
41.6 |
12 |
PDB header:chaperone
Chain: B: PDB Molecule:endoplasmin;
PDBTitle: structure of middle plus c-terminal domains (m+c) of grp94
|
| 75 | c2vzkD_ |
|
not modelled |
41.3 |
19 |
PDB header:transferase
Chain: D: PDB Molecule:glutamate n-acetyltransferase 2 beta chain;
PDBTitle: structure of the acyl-enzyme complex of an n-terminal2 nucleophile (ntn) hydrolase, oat2
|
| 76 | c2qw5B_ |
|
not modelled |
40.0 |
9 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase-like tim barrel;
PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
|
| 77 | c3m1rF_ |
|
not modelled |
39.8 |
32 |
PDB header:hydrolase
Chain: F: PDB Molecule:formimidoylglutamase;
PDBTitle: the crystal structure of formimidoylglutamase from bacillus2 subtilis subsp. subtilis str. 168
|
| 78 | c3no4A_ |
|
not modelled |
39.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:creatinine amidohydrolase;
PDBTitle: crystal structure of a creatinine amidohydrolase (npun_f1913) from2 nostoc punctiforme pcc 73102 at 2.00 a resolution
|
| 79 | c3o1lB_ |
|
not modelled |
39.0 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
|
| 80 | c2gq0B_ |
|
not modelled |
38.1 |
19 |
PDB header:chaperone, hydrolase
Chain: B: PDB Molecule:chaperone protein htpg;
PDBTitle: crystal structure of the middle domain of htpg, the e. coli2 hsp90
|
| 81 | c1y6zA_ |
|
not modelled |
36.8 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:heat shock protein, putative;
PDBTitle: middle domain of plasmodium falciparum putative heat shock protein2 pf14_0417
|
| 82 | c3dxiB_ |
|
not modelled |
36.3 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative aldolase;
PDBTitle: crystal structure of the n-terminal domain of a putative2 aldolase (bvu_2661) from bacteroides vulgatus
|
| 83 | c3tr9A_ |
|
not modelled |
35.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dihydropteroate synthase;
PDBTitle: structure of a dihydropteroate synthase (folp) in complex with pteroic2 acid from coxiella burnetii
|
| 84 | c3nrbD_ |
|
not modelled |
34.0 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
| 85 | c3mmrA_ |
|
not modelled |
33.6 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:arginase;
PDBTitle: structure of plasmodium falciparum arginase in complex with abh
|
| 86 | c1nofA_ |
|
not modelled |
33.3 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:xylanase;
PDBTitle: the first crystallographic structure of a xylanase from2 glycosyl hydrolase family 5: implications for catalysis
|
| 87 | c3icgD_ |
|
not modelled |
33.3 |
16 |
PDB header:hydrolase
Chain: D: PDB Molecule:endoglucanase d;
PDBTitle: crystal structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
|
| 88 | c3pzlA_ |
|
not modelled |
32.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:agmatine ureohydrolase;
PDBTitle: the crystal structure of agmatine ureohydrolase of thermoplasma2 volcanium
|
| 89 | c3obiC_ |
|
not modelled |
31.9 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
| 90 | d1vjia_ |
|
not modelled |
31.9 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
| 91 | d2aeba1 |
|
not modelled |
31.7 |
19 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 92 | d1luaa2 |
|
not modelled |
31.1 |
22 |
Fold:Aminoacid dehydrogenase-like, N-terminal domain
Superfamily:Aminoacid dehydrogenase-like, N-terminal domain
Family:Methylene-tetrahydromethanopterin dehydrogenase |
| 93 | c3i05B_ |
|
not modelled |
30.4 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: tryptophanyl-trna synthetase from trypanosoma brucei
|
| 94 | c2cgeD_ |
|
not modelled |
30.3 |
14 |
PDB header:chaperone
Chain: D: PDB Molecule:atp-dependent molecular chaperone hsp82;
PDBTitle: crystal structure of an hsp90-sba1 closed chaperone complex
|
| 95 | c1gqkB_ |
|
not modelled |
30.2 |
55 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-d-glucuronidase;
PDBTitle: structure of pseudomonas cellulosa alpha-d-glucuronidase2 complexed with glucuronic acid
|
| 96 | c1ydnA_ |
|
not modelled |
29.9 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of the hmg-coa lyase from brucella melitensis,2 northeast structural genomics target lr35.
|
| 97 | c1wr2A_ |
|
not modelled |
29.9 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ph1789;
PDBTitle: crystal structure of ph1788 from pyrococcus horikoshii ot3
|
| 98 | c3bleA_ |
|
not modelled |
29.3 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in2 complexed with malonate
|
| 99 | d1d3va_ |
|
not modelled |
28.2 |
18 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
| 100 | d1oy0a_ |
|
not modelled |
28.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
| 101 | d1zs9a1 |
|
not modelled |
27.8 |
13 |
Fold:HAD-like
Superfamily:HAD-like
Family:Enolase-phosphatase E1 |
| 102 | c3ih1A_ |
|
not modelled |
27.7 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 103 | c3n2tA_ |
|
not modelled |
27.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: structure of the glycerol dehydrogenase akr11b4 from gluconobacter2 oxydans
|
| 104 | c3lyeA_ |
|
not modelled |
27.2 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
| 105 | c3epmB_ |
|
not modelled |
27.1 |
19 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:thiamine biosynthesis protein thic;
PDBTitle: crystal structure of caulobacter crescentus thic
|
| 106 | c3q6nF_ |
|
not modelled |
27.0 |
17 |
PDB header:chaperone
Chain: F: PDB Molecule:heat shock protein hsp 90-alpha;
PDBTitle: crystal structure of human mc-hsp90 in p21 space group
|
| 107 | d2p02a1 |
|
not modelled |
26.9 |
20 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 108 | c3opbA_ |
|
not modelled |
24.9 |
19 |
PDB header:protein binding
Chain: A: PDB Molecule:swi5-dependent ho expression protein 4;
PDBTitle: crystal structure of she4p
|
| 109 | d2hcfa1 |
|
not modelled |
24.8 |
9 |
Fold:HAD-like
Superfamily:HAD-like
Family:beta-Phosphoglucomutase-like |
| 110 | c3mwdB_ |
|
not modelled |
24.8 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:atp-citrate synthase;
PDBTitle: truncated human atp-citrate lyase with citrate bound
|
| 111 | c3krsB_ |
|
not modelled |
24.8 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: structure of triosephosphate isomerase from cryptosporidium parvum at2 1.55a resolution
|
| 112 | d1r5ja_ |
|
not modelled |
24.6 |
14 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 113 | c1vm6B_ |
|
not modelled |
24.5 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: crystal structure of dihydrodipicolinate reductase (tm1520) from2 thermotoga maritima at 2.27 a resolution
|
| 114 | d1qh8b_ |
|
not modelled |
24.3 |
19 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
| 115 | c3pryA_ |
|
not modelled |
24.2 |
14 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein hsp 90-beta;
PDBTitle: crystal structure of the middle domain of human hsp90-beta refined at2 2.3 a resolution
|
| 116 | c3gkaB_ |
|
not modelled |
23.7 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
| 117 | d1rqba2 |
|
not modelled |
23.4 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
| 118 | c3hjcA_ |
|
not modelled |
23.0 |
24 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein 83-1;
PDBTitle: crystal structure of the carboxy-terminal domain of hsp90 from2 leishmania major, lmjf33.0312
|
| 119 | c2v1dA_ |
|
not modelled |
22.7 |
15 |
PDB header:oxidoreductase/repressor
Chain: A: PDB Molecule:lysine-specific histone demethylase 1;
PDBTitle: structural basis of lsd1-corest selectivity in histone h32 recognition
|
| 120 | c3cyvA_ |
|
not modelled |
22.3 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 shigella flexineri: new insights into its catalytic3 mechanism
|