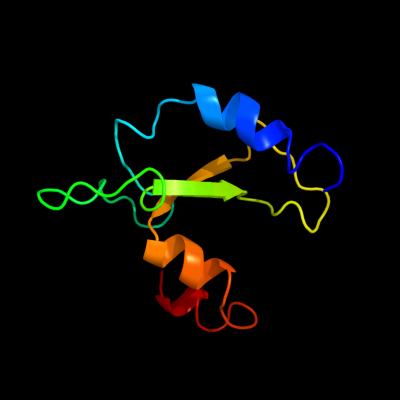
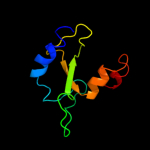
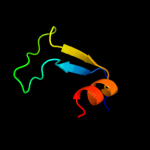
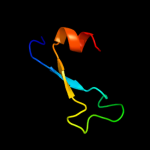
1 c2qgpA_
98.9
26
PDB header: hydrolaseChain: A: PDB Molecule: hnh endonuclease;PDBTitle: x-ray structure of the nhn endonuclease from geobacter2 metallireducens. northeast structural genomics consortium3 target gmr87.
2 d2gykb1
96.1
34
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: HNH-motif3 c7ceiB_
88.4
31
PDB header: immune systemChain: B: PDB Molecule: protein (colicin e7 immunity protein);PDBTitle: the endonuclease domain of colicin e7 in complex with its inhibitor2 im7 protein
4 c2qnfB_
82.7
20
PDB header: hydrolase/dnaChain: B: PDB Molecule: recombination endonuclease vii;PDBTitle: crystal structure of t4 endonuclease vii h43n mutant in2 complex with heteroduplex dna containing base mismatches
5 d1e7la2
80.3
22
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: Recombination endonuclease VII, N-terminal domain6 c2orvB_
76.1
32
PDB header: transferaseChain: B: PDB Molecule: thymidine kinase;PDBTitle: human thymidine kinase 1 in complex with tp4a
7 c1w4rC_
72.2
30
PDB header: transferaseChain: C: PDB Molecule: thymidine kinase;PDBTitle: structure of a type ii thymidine kinase with bound dttp
8 c2b8tA_
69.9
26
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: crystal structure of thymidine kinase from u.urealyticum in2 complex with thymidine
9 c2qq0B_
66.5
32
PDB header: transferaseChain: B: PDB Molecule: thymidine kinase;PDBTitle: thymidine kinase from thermotoga maritima in complex with2 thymidine + appnhp
10 d2jb0b1
63.0
32
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: HNH-motif11 d2b8ta2
59.5
22
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Type II thymidine kinase zinc finger12 d1xbta2
50.8
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Type II thymidine kinase zinc finger13 c3goxB_
44.7
20
PDB header: hydrolase/dnaChain: B: PDB Molecule: restriction endonuclease hpy99i;PDBTitle: crystal structure of the beta-beta-alpha-me type ii restriction2 endonuclease hpy99i in the absence of edta
14 d1jhfa1
42.6
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LexA repressor, N-terminal DNA-binding domain15 c1xx6B_
40.5
24
PDB header: transferaseChain: B: PDB Molecule: thymidine kinase;PDBTitle: x-ray structure of clostridium acetobutylicum thymidine kinase with2 adp. northeast structural genomics target car26.
16 c2iqjB_
38.4
31
PDB header: protein transportChain: B: PDB Molecule: stromal membrane-associated protein 1-like;PDBTitle: crystal structure of the gap domain of smap1l (loc64744)2 stromal membrane-associated protein 1-like
17 c3e2iA_
37.7
20
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: crystal structure of thymidine kinase from s. aureus
18 c2vr0A_
34.0
20
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c nitrite reductase, catalytic subunit nfra;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex bound to the hqno inhibitor
19 d1qdba_
29.4
24
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif20 c1tvtA_
27.3
22
PDB header: transcription regulationChain: A: PDB Molecule: transactivator protein;PDBTitle: structure of the equine infectious anemia virus tat protein
21 c3h7hA_
not modelled
24.2
26
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt4;PDBTitle: crystal structure of the human transcription elongation factor dsif,2 hspt4/hspt5 (176-273)
22 d1vp8a_
not modelled
23.8
28
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: MTH1675-like23 d2dj7a1
not modelled
23.3
33
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain24 c3lvrE_
not modelled
23.1
36
PDB header: protein transportChain: E: PDB Molecule: arf-gap with sh3 domain, ank repeat and ph domain-PDBTitle: the crystal structure of asap3 in complex with arf6 in transition2 state soaked with calcium
25 c2ju5A_
not modelled
22.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: thioredoxin disulfide isomerase;PDBTitle: dsbh oxidoreductase
26 c2p57A_
not modelled
21.7
29
PDB header: metal binding proteinChain: A: PDB Molecule: gtpase-activating protein znf289;PDBTitle: gap domain of znf289, an id1-regulated zinc finger protein
27 d2glia5
not modelled
21.5
46
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H228 d1jx6a_
not modelled
21.5
22
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like29 c2wy3B_
not modelled
21.0
48
PDB header: immune system/viral proteinChain: B: PDB Molecule: uncharacterized protein ul16;PDBTitle: structure of the hcmv ul16-micb complex elucidates select2 binding of a viral immunoevasin to diverse nkg2d ligands
30 c2f3oB_
not modelled
20.2
24
PDB header: unknown functionChain: B: PDB Molecule: pyruvate formate-lyase 2;PDBTitle: crystal structure of a glycyl radical enzyme from archaeoglobus2 fulgidus
31 d1xx6a2
not modelled
19.8
29
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: Type II thymidine kinase zinc finger32 c3hcwB_
not modelled
19.8
25
PDB header: rna binding proteinChain: B: PDB Molecule: maltose operon transcriptional repressor;PDBTitle: crystal structure of probable maltose operon transcriptional repressor2 malr from staphylococcus areus
33 c2ja1A_
not modelled
19.3
21
PDB header: transferaseChain: A: PDB Molecule: thymidine kinase;PDBTitle: thymidine kinase from b. cereus with ttp bound as phosphate2 donor.
34 c3m7kA_
not modelled
18.9
22
PDB header: hydrolase/dnaChain: A: PDB Molecule: restriction endonuclease paci;PDBTitle: crystal structure of paci-dna enzyme product complex
35 c3na7A_
not modelled
17.9
34
PDB header: gene regulation, chaperoneChain: A: PDB Molecule: hp0958;PDBTitle: 2.2 angstrom structure of the hp0958 protein from helicobacter pylori2 ccug 17874
36 d1x3ha2
not modelled
17.7
35
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain37 d2jnea1
not modelled
17.5
54
Fold: Rubredoxin-likeSuperfamily: YfgJ-likeFamily: YfgJ-like38 c2jneA_
not modelled
17.5
54
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein yfgj;PDBTitle: nmr structure of e.coli yfgj modelled with two zn+2 bound.2 northeast structural genomics consortium target er317.
39 c3iraA_
not modelled
17.1
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein;PDBTitle: the crystal structure of one domain of the conserved protein from2 methanosarcina mazei go1
40 c1yshD_
not modelled
16.9
53
PDB header: structural protein/rnaChain: D: PDB Molecule: ribosomal protein l37a;PDBTitle: localization and dynamic behavior of ribosomal protein l30e
41 d1jj2y_
not modelled
16.5
26
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae42 d1r9da_
not modelled
16.1
22
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: PFL-like43 c2x41A_
not modelled
16.1
42
PDB header: hydrolaseChain: A: PDB Molecule: beta-glucosidase;PDBTitle: structure of beta-glucosidase 3b from thermotoga neapolitana2 in complex with glucose
44 c2d9lA_
not modelled
16.0
24
PDB header: gene regulationChain: A: PDB Molecule: nucleoporin-like protein rip;PDBTitle: solution structure of the arfgap domain of human rip
45 c2zkrz_
not modelled
16.0
41
PDB header: ribosomal protein/rnaChain: Z: PDB Molecule: e site t-rna;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
46 c2j7aE_
not modelled
15.3
18
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c nitrite reductase nrfa;PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex from desulfovibrio vulgaris
47 c2jrpA_
not modelled
15.2
46
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative cytoplasmic protein;PDBTitle: solution nmr structure of yfgj from salmonella typhimurium2 modeled with two zn+2 bound, northeast structural genomics3 consortium target str86
48 c3dwdB_
not modelled
15.0
31
PDB header: transport proteinChain: B: PDB Molecule: adp-ribosylation factor gtpase-activating protein 1;PDBTitle: crystal structure of the arfgap domain of human arfgap1
49 d1vqoz1
not modelled
14.8
21
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae50 c1fs9A_
not modelled
14.7
33
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c nitrite reductase;PDBTitle: cytochrome c nitrite reductase from wolinella succinogenes-azide2 complex
51 c2b0oF_
not modelled
14.5
38
PDB header: metal binding proteinChain: F: PDB Molecule: uplc1;PDBTitle: crystal structure of uplc1 gap domain
52 c2joxA_
not modelled
14.1
28
PDB header: transcriptionChain: A: PDB Molecule: churchill protein;PDBTitle: embryonic neural inducing factor churchill is not a dna-2 binding zinc finger protein: solution structure reveals a3 solvent-exposed beta-sheet and zinc binuclear cluster
53 c1i6hA_
not modelled
14.0
17
PDB header: transcription/dna-rna hybridChain: A: PDB Molecule: dna-directed rna polymerase ii largest subunit;PDBTitle: rna polymerase ii elongation complex
54 d2ic1a1
not modelled
13.9
26
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Cysteine dioxygenase type I55 c3qqcA_
not modelled
13.8
21
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase subunit b, dna-directed rnaPDBTitle: crystal structure of archaeal spt4/5 bound to the rnap clamp domain
56 d3elna1
not modelled
13.6
26
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Cysteine dioxygenase type I57 c2y8nC_
not modelled
13.5
20
PDB header: lyaseChain: C: PDB Molecule: 4-hydroxyphenylacetate decarboxylase large subunit;PDBTitle: crystal structure of glycyl radical enzyme
58 d1fs7a_
not modelled
13.3
33
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Di-heme elbow motif59 c3cc4Z_
not modelled
13.3
24
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein l37ae;PDBTitle: co-crystal structure of anisomycin bound to the 50s ribosomal subunit
60 c4a17Y_
not modelled
13.2
47
PDB header: ribosomeChain: Y: PDB Molecule: rpl37a;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
61 d1vh3a_
not modelled
12.8
21
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: Cytidylytransferase62 d1ubdc1
not modelled
12.6
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H263 d1weoa_
not modelled
12.5
21
Fold: RING/U-boxSuperfamily: RING/U-boxFamily: RING finger domain, C3HC464 d1ffkw_
not modelled
12.4
19
Fold: Rubredoxin-likeSuperfamily: Zn-binding ribosomal proteinsFamily: Ribosomal protein L37ae65 d1inza_
not modelled
11.6
31
Fold: alpha-alpha superhelixSuperfamily: ENTH/VHS domainFamily: ENTH domain66 c3f9uA_
not modelled
11.1
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative exported cytochrome c biogenesis-related protein;PDBTitle: crystal structure of c-terminal domain of putative exported cytochrome2 c biogenesis-related protein from bacteroides fragilis
67 d1twfa_
not modelled
10.9
17
Fold: beta and beta-prime subunits of DNA dependent RNA-polymeraseSuperfamily: beta and beta-prime subunits of DNA dependent RNA-polymeraseFamily: RNA-polymerase beta-prime68 c3jyw9_
not modelled
10.9
29
PDB header: ribosomeChain: 9: PDB Molecule: 60s ribosomal protein l43;PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
69 c1rikA_
not modelled
10.6
38
PDB header: de novo proteinChain: A: PDB Molecule: e6apc1 peptide;PDBTitle: e6-binding zinc finger (e6apc1)
70 d1hs5a_
not modelled
10.6
100
Fold: p53 tetramerization domainSuperfamily: p53 tetramerization domainFamily: p53 tetramerization domain71 d1n5ga_
not modelled
10.5
33
Fold: Zinc finger domain of DNA polymerase-alphaSuperfamily: Zinc finger domain of DNA polymerase-alphaFamily: Zinc finger domain of DNA polymerase-alpha72 d2fiya1
not modelled
10.4
32
Fold: FdhE-likeSuperfamily: FdhE-likeFamily: FdhE-like73 c1x4lA_
not modelled
10.2
28
PDB header: metal binding proteinChain: A: PDB Molecule: skeletal muscle lim-protein 3;PDBTitle: solution structure of lim domain in four and a half lim2 domains protein 2
74 c3cueH_
not modelled
9.9
25
PDB header: protein transportChain: H: PDB Molecule: transport protein particle 31 kda subunit;PDBTitle: crystal structure of a trapp subassembly activating the rab2 ypt1p
75 c2qu7B_
not modelled
9.8
21
PDB header: transcriptionChain: B: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of a putative transcription regulator2 from staphylococcus saprophyticus subsp. saprophyticus
76 d1h16a_
not modelled
9.6
20
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: PFL-like77 c2l4zA_
not modelled
9.5
26
PDB header: hydrolase, metal binding proteinChain: A: PDB Molecule: dna endonuclease rbbp8, lim domain transcription factorPDBTitle: nmr structure of fusion of ctip (641-685) to lmo4-lim1 (18-82)
78 c3pnnA_
not modelled
9.2
22
PDB header: transferaseChain: A: PDB Molecule: conserved domain protein;PDBTitle: the crystal structure of a glycosyltransferase from porphyromonas2 gingivalis w83
79 c3c3kA_
not modelled
9.1
20
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: crystal structure of an uncharacterized protein from actinobacillus2 succinogenes
80 c3gbvB_
not modelled
9.1
13
PDB header: transcription regulatorChain: B: PDB Molecule: putative laci-family transcriptional regulator;PDBTitle: crystal structure of a putative laci transcriptional regulator from2 bacteroides fragilis
81 c1m3vA_
not modelled
9.1
15
PDB header: metal binding proteinChain: A: PDB Molecule: fusion of the lim interacting domain of ldb1 andPDBTitle: flin4: fusion of the lim binding domain of ldb1 and the n-2 terminal lim domain of lmo4
82 c3o47A_
not modelled
9.1
28
PDB header: hydrolase, hydrolase activatorChain: A: PDB Molecule: adp-ribosylation factor gtpase-activating protein 1, adp-PDBTitle: crystal structure of arfgap1-arf1 fusion protein
83 c3jywY_
not modelled
9.0
38
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l37(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
84 c2qh8A_
not modelled
8.9
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of conserved domain protein from vibrio2 cholerae o1 biovar eltor str. n16961
85 c1b8tA_
not modelled
8.9
32
PDB header: contractileChain: A: PDB Molecule: protein (crp1);PDBTitle: solution structure of the chicken crp1
86 c1vhkA_
not modelled
8.8
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yqeu;PDBTitle: crystal structure of an hypothetical protein
87 c3fxhA_
not modelled
8.8
58
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: integron gene cassette protein hfx_cass2;PDBTitle: crystal structure from the mobile metagenome of halifax2 harbour sewage outfall: integron cassette protein hfx_cass2
88 c2zkr2_
not modelled
8.7
38
PDB header: ribosomal protein/rnaChain: 2: PDB Molecule: 60s ribosomal protein l37e;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
89 c2r5iL_
not modelled
8.7
33
PDB header: viral proteinChain: L: PDB Molecule: l1 protein;PDBTitle: pentamer structure of major capsid protein l1 of human2 papilloma virus type 18
90 c2o20H_
not modelled
8.5
30
PDB header: transcriptionChain: H: PDB Molecule: catabolite control protein a;PDBTitle: crystal structure of transcription regulator ccpa of lactococcus2 lactis
91 c3ac0B_
not modelled
8.4
35
PDB header: hydrolaseChain: B: PDB Molecule: beta-glucosidase i;PDBTitle: crystal structure of beta-glucosidase from kluyveromyces marxianus in2 complex with glucose
92 c1s1iY_
not modelled
8.3
38
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l37-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
93 d1qcva_
not modelled
8.3
20
Fold: Rubredoxin-likeSuperfamily: Rubredoxin-likeFamily: Rubredoxin94 c3gn5B_
not modelled
8.3
15
PDB header: dna binding proteinChain: B: PDB Molecule: hth-type transcriptional regulator mqsa (ygit/b3021);PDBTitle: structure of the e. coli protein mqsa (ygit/b3021)
95 c3mizB_
not modelled
8.1
14
PDB header: transcription regulatorChain: B: PDB Molecule: putative transcriptional regulator protein, laciPDBTitle: crystal structure of a putative transcriptional regulator2 protein, lacl family from rhizobium etli
96 d2yt9a2
not modelled
8.0
83
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H297 c3e56A_
not modelled
8.0
19
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: the 2.0 angstrom resolution crystal structure of npr1517, a putative2 heterocyst differentiation inhibitor from nostoc punctiforme
98 d2dlqa4
not modelled
7.8
67
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H299 c1tjlD_
not modelled
7.8
56
PDB header: transcriptionChain: D: PDB Molecule: dnak suppressor protein;PDBTitle: crystal structure of transcription factor dksa from e. coli