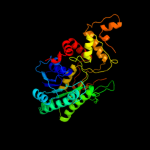
| 1 | d1bxka_ |
|
 |
100.0 |
97 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
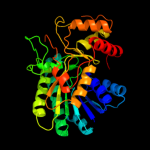
| 2 | c1z45A_ |
|
 |
100.0 |
27 |
PDB header:isomerase
Chain: A: PDB Molecule:gal10 bifunctional protein;
PDBTitle: crystal structure of the gal10 fusion protein galactose2 mutarotase/udp-galactose 4-epimerase from saccharomyces3 cerevisiae complexed with nad, udp-glucose, and galactose
|
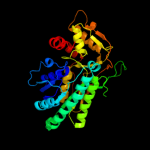
| 3 | d1kewa_ |
|
 |
100.0 |
75 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
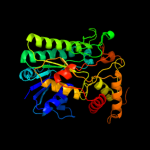
| 4 | c1z7eC_ |
|
 |
100.0 |
19 |
PDB header:hydrolase
Chain: C: PDB Molecule:protein arna;
PDBTitle: crystal structure of full length arna
|
| 5 | d1oc2a_ |
|
 |
100.0 |
42 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 6 | d1r6da_ |
|
 |
100.0 |
46 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 7 | d1i24a_ |
|
 |
100.0 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 8 | c2hunB_ |
|
 |
100.0 |
47 |
PDB header:lyase
Chain: B: PDB Molecule:336aa long hypothetical dtdp-glucose 4,6-dehydratase;
PDBTitle: crystal structure of hypothetical protein ph0414 from pyrococcus2 horikoshii ot3
|
| 9 | c1n7gB_ |
|
 |
100.0 |
24 |
PDB header:lyase
Chain: B: PDB Molecule:gdp-d-mannose-4,6-dehydratase;
PDBTitle: crystal structure of the gdp-mannose 4,6-dehydratase2 ternary complex with nadph and gdp-rhamnose.
|
| 10 | d1n7ha_ |
|
 |
100.0 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 11 | c2pk3B_ |
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-6-deoxy-d-lyxo-4-hexulose reductase;
PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
|
| 12 | d1z45a2 |
|
 |
100.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 13 | d1ek6a_ |
|
 |
100.0 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 14 | c2z1mC_ |
|
 |
100.0 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:gdp-d-mannose dehydratase;
PDBTitle: crystal structure of gdp-d-mannose dehydratase from aquifex aeolicus2 vf5
|
| 15 | c3enkB_ |
|
 |
100.0 |
22 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: 1.9a crystal structure of udp-glucose 4-epimerase from2 burkholderia pseudomallei
|
| 16 | d1t2aa_ |
|
 |
100.0 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 17 | d1udca_ |
|
 |
100.0 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 18 | c1t2aC_ |
|
 |
100.0 |
19 |
PDB header:structural genomics,lyase
Chain: C: PDB Molecule:gdp-mannose 4,6 dehydratase;
PDBTitle: crystal structure of human gdp-d-mannose 4,6-dehydratase
|
| 19 | d1db3a_ |
|
 |
100.0 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 20 | d1wvga1 |
|
 |
100.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 21 | c2q1uA_ |
|
not modelled |
100.0 |
26 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmf in2 complex with nad+ and udp
|
| 22 | d1rpna_ |
|
not modelled |
100.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 23 | d1rkxa_ |
|
not modelled |
100.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 24 | d2c5aa1 |
|
not modelled |
100.0 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 25 | c2c20D_ |
|
not modelled |
100.0 |
27 |
PDB header:isomerase
Chain: D: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of udp-glucose 4-epimerase
|
| 26 | d2blla1 |
|
not modelled |
100.0 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 27 | c2pzlB_ |
|
not modelled |
100.0 |
20 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme2 wbmg in complex with nad and udp
|
| 28 | c2p5uC_ |
|
not modelled |
100.0 |
28 |
PDB header:isomerase
Chain: C: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of thermus thermophilus hb8 udp-glucose 4-2 epimerase complex with nad
|
| 29 | c2b69A_ |
|
not modelled |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:udp-glucuronate decarboxylase 1;
PDBTitle: crystal structure of human udp-glucoronic acid decarboxylase
|
| 30 | d2b69a1 |
|
not modelled |
100.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 31 | c2q1wC_ |
|
not modelled |
100.0 |
21 |
PDB header:sugar binding protein
Chain: C: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmh in2 complex with nad+
|
| 32 | d1orra_ |
|
not modelled |
100.0 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 33 | c3slgB_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:pbgp3 protein;
PDBTitle: crystal structure of pbgp3 protein from burkholderia pseudomallei
|
| 34 | d1sb8a_ |
|
not modelled |
100.0 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 35 | c2iodD_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:dihydroflavonol 4-reductase;
PDBTitle: binding of two substrate analogue molecules to2 dihydroflavonol-4-reductase alters the functional geometry3 of the catalytic site
|
| 36 | d1gy8a_ |
|
not modelled |
100.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 37 | d1e6ua_ |
|
not modelled |
100.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 38 | c3lu1C_ |
|
not modelled |
100.0 |
25 |
PDB header:isomerase
Chain: C: PDB Molecule:wbgu;
PDBTitle: crystal structure analysis of wbgu: a udp-galnac 4-epimerase
|
| 39 | c3m2pD_ |
|
not modelled |
100.0 |
19 |
PDB header:isomerase
Chain: D: PDB Molecule:udp-n-acetylglucosamine 4-epimerase;
PDBTitle: the crystal structure of udp-n-acetylglucosamine 4-epimerase2 from bacillus cereus
|
| 40 | c2x4gA_ |
|
not modelled |
100.0 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate-sugar epimerase;
PDBTitle: crystal structure of pa4631, a nucleoside-diphosphate-sugar2 epimerase from pseudomonas aeruginosa
|
| 41 | c2p4hX_ |
|
not modelled |
100.0 |
17 |
PDB header:plant protein
Chain: X: PDB Molecule:vestitone reductase;
PDBTitle: crystal structure of vestitone reductase from alfalfa2 (medicago sativa l.)
|
| 42 | c2hrzA_ |
|
not modelled |
100.0 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nucleoside-diphosphate-sugar epimerase;
PDBTitle: the crystal structure of the nucleoside-diphosphate-sugar epimerase2 from agrobacterium tumefaciens
|
| 43 | c3a1nB_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ndp-sugar epimerase;
PDBTitle: crystal structure of l-threonine dehydrogenase from2 hyperthermophilic archaeon thermoplasma volcanium
|
| 44 | c3eheB_ |
|
not modelled |
100.0 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-glucose 4-epimerase (gale-1);
PDBTitle: crystal structure of udp-glucose 4 epimerase (gale-1) from2 archaeoglobus fulgidus
|
| 45 | c2yy7B_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-threonine dehydrogenase;
PDBTitle: crystal structure of thermolabile l-threonine dehydrogenase from2 flavobacterium frigidimaris kuc-1
|
| 46 | c2rh8A_ |
|
not modelled |
100.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:anthocyanidin reductase;
PDBTitle: structure of apo anthocyanidin reductase from vitis vinifera
|
| 47 | c2v6gA_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:progesterone 5-beta-reductase;
PDBTitle: structure of progesterone 5beta-reductase from digitalis2 lanata in complex with nadp
|
| 48 | c3icpA_ |
|
not modelled |
100.0 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of udp-galactose 4-epimerase
|
| 49 | d1n2sa_ |
|
not modelled |
100.0 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 50 | d1vl0a_ |
|
not modelled |
100.0 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 51 | c3sc6F_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: 2.65 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 reductase (rfbd) from bacillus anthracis str. ames in complex with3 nadp
|
| 52 | d1y1pa1 |
|
not modelled |
100.0 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 53 | c2x86K_ |
|
not modelled |
100.0 |
17 |
PDB header:isomerase
Chain: K: PDB Molecule:adp-l-glycero-d-manno-heptose-6-epimerase;
PDBTitle: agme bound to adp-b-mannose
|
| 54 | c2ydyA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine adenosyltransferase 2 subunit beta;
PDBTitle: crystal structure of human s-adenosylmethionine synthetase2 2, beta subunit in orthorhombic crystal form
|
| 55 | d1eq2a_ |
|
not modelled |
100.0 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 56 | c3gpiA_ |
|
not modelled |
100.0 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: structure of putative nad-dependent epimerase/dehydratase2 from methylobacillus flagellatus
|
| 57 | c3oh8A_ |
|
not modelled |
100.0 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate sugar epimerase (sula family);
PDBTitle: crystal structure of the nucleoside-diphosphate sugar epimerase from2 corynebacterium glutamicum. northeast structural genomics consortium3 target cgr91
|
| 58 | c2ggsB_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:273aa long hypothetical dtdp-4-dehydrorhamnose
PDBTitle: crystal structure of hypothetical dtdp-4-dehydrorhamnose2 reductase from sulfolobus tokodaii
|
| 59 | c3iusB_ |
|
not modelled |
100.0 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized conserved protein;
PDBTitle: the structure of a functionally unknown conserved protein2 from silicibacter pomeroyi dss
|
| 60 | c2gn9B_ |
|
not modelled |
99.9 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:udp-glcnac c6 dehydratase;
PDBTitle: crystal structure of udp-glcnac inverting 4,6-dehydratase in complex2 with nadp and udp-glc
|
| 61 | c3e48B_ |
|
not modelled |
99.9 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative nucleoside-diphosphate-sugar epimerase;
PDBTitle: crystal structure of a nucleoside-diphosphate-sugar epimerase2 (sav0421) from staphylococcus aureus, northeast structural genomics3 consortium target zr319
|
| 62 | d1xgka_ |
|
not modelled |
99.9 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 63 | c2qx7A_ |
|
not modelled |
99.9 |
15 |
PDB header:plant protein
Chain: A: PDB Molecule:eugenol synthase 1;
PDBTitle: structure of eugenol synthase from ocimum basilicum
|
| 64 | c2zklA_ |
|
not modelled |
99.9 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:capsular polysaccharide synthesis enzyme cap5f;
PDBTitle: crystal structure of capsular polysaccharide assembling protein capf2 from staphylococcus aureus
|
| 65 | c3i5mA_ |
|
not modelled |
99.9 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative leucoanthocyanidin reductase 1;
PDBTitle: structure of the apo form of leucoanthocyanidin reductase from vitis2 vinifera
|
| 66 | c3c1oA_ |
|
not modelled |
99.9 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:eugenol synthase;
PDBTitle: the multiple phenylpropene synthases in both clarkia2 breweri and petunia hybrida represent two distinct lineages
|
| 67 | d1qyda_ |
|
not modelled |
99.9 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 68 | c2zcuA_ |
|
not modelled |
99.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized oxidoreductase ytfg;
PDBTitle: crystal structure of a new type of nadph-dependent quinone2 oxidoreductase (qor2) from escherichia coli
|
| 69 | c2vrcD_ |
|
not modelled |
99.9 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:triphenylmethane reductase;
PDBTitle: crystal structure of the citrobacter sp. triphenylmethane2 reductase complexed with nadp(h)
|
| 70 | d1qyca_ |
|
not modelled |
99.9 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 71 | c2exxB_ |
|
not modelled |
99.9 |
12 |
PDB header:unknown function
Chain: B: PDB Molecule:hscarg protein;
PDBTitle: crystal structure of hscarg from homo sapiens in complex with nadp
|
| 72 | c2gasA_ |
|
not modelled |
99.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isoflavone reductase;
PDBTitle: crystal structure of isoflavone reductase
|
| 73 | c3nzoB_ |
|
not modelled |
99.8 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:udp-n-acetylglucosamine 4,6-dehydratase;
PDBTitle: udp-n-acetylglucosamine 4,6-dehydratase from vibrio fischeri.
|
| 74 | c3ay3C_ |
|
not modelled |
99.8 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of glucuronic acid dehydrogeanse from2 chromohalobacter salexigens
|
| 75 | c3rfxB_ |
|
not modelled |
99.8 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uronate dehydrogenase;
PDBTitle: crystal structure of uronate dehydrogenase from agrobacterium2 tumefaciens, y136a mutant complexed with nad
|
| 76 | d2q46a1 |
|
not modelled |
99.8 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 77 | d1hdoa_ |
|
not modelled |
99.7 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 78 | c3dhnA_ |
|
not modelled |
99.7 |
18 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of the putative epimerase q89z24_bactn2 from bacteroides thetaiotaomicron. northeast structural3 genomics consortium target btr310.
|
| 79 | c3e8xA_ |
|
not modelled |
99.7 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative nad-dependent epimerase/dehydratase;
PDBTitle: putative nad-dependent epimerase/dehydratase from bacillus halodurans.
|
| 80 | c3dqpA_ |
|
not modelled |
99.7 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase ylbe;
PDBTitle: crystal structure of the oxidoreductase ylbe from2 lactococcus lactis, northeast structural genomics3 consortium target kr121.
|
| 81 | c3h2sA_ |
|
not modelled |
99.6 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative nadh-flavin reductase;
PDBTitle: crystal structure of the q03b84 protein from lactobacillus2 casei. northeast structural genomics consortium target3 lcr19.
|
| 82 | c3qvoA_ |
|
not modelled |
99.6 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nmra family protein;
PDBTitle: structure of a rossmann-fold nad(p)-binding family protein from2 shigella flexneri.
|
| 83 | d2bkaa1 |
|
not modelled |
99.5 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 84 | d2a35a1 |
|
not modelled |
99.4 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 85 | c3ew7A_ |
|
not modelled |
99.4 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lmo0794 protein;
PDBTitle: crystal structure of the lmo0794 protein from listeria2 monocytogenes. northeast structural genomics consortium3 target lmr162.
|
| 86 | c2jahB_ |
|
not modelled |
99.3 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:clavulanic acid dehydrogenase;
PDBTitle: biochemical and structural analysis of the clavulanic acid2 dehydeogenase (cad) from streptomyces clavuligerus
|
| 87 | c3l77A_ |
|
not modelled |
99.3 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain alcohol dehydrogenase;
PDBTitle: x-ray structure alcohol dehydrogenase from archaeon thermococcus2 sibiricus complexed with 5-hydroxy-nadp
|
| 88 | d2gdza1 |
|
not modelled |
99.2 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 89 | d2fmua1 |
|
not modelled |
99.2 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 90 | c2qioA_ |
|
not modelled |
99.2 |
11 |
PDB header:unknown function
Chain: A: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: x-ray structure of enoyl-acyl carrier protein reductase from bacillus2 anthracis with triclosan
|
| 91 | c3i4fD_ |
|
not modelled |
99.2 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-oxoacyl-[acyl-carrier protein] reductase;
PDBTitle: structure of putative 3-oxoacyl-reductase from bacillus thuringiensis
|
| 92 | c2p68A_ |
|
not modelled |
99.2 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of aq_1716 from aquifex aeolicus vf5
|
| 93 | c3v8bC_ |
|
not modelled |
99.2 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dehydrogenase, possibly 3-oxoacyl-[acyl-carrier
PDBTitle: crystal structure of a 3-ketoacyl-acp reductase from sinorhizobium2 meliloti 1021
|
| 94 | c3oidA_ |
|
not modelled |
99.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadph];
PDBTitle: crystal structure of enoyl-acp reductases iii (fabl) from b. subtilis2 (complex with nadp and tcl)
|
| 95 | c3m1aF_ |
|
not modelled |
99.1 |
14 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative dehydrogenase;
PDBTitle: the crystal structure of a short-chain dehydrogenase from2 streptomyces avermitilis to 2a
|
| 96 | c2cfcB_ |
|
not modelled |
99.1 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-(r)-hydroxypropyl-com dehydrogenase;
PDBTitle: structural basis for stereo selectivity in the (r)- and2 (s)-hydroxypropylethane thiosulfonate dehydrogenases
|
| 97 | c3p19A_ |
|
not modelled |
99.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative blue fluorescent protein;
PDBTitle: improved nadph-dependent blue fluorescent protein
|
| 98 | c3toxG_ |
|
not modelled |
99.1 |
14 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase in complex with2 nad(p) from sinorhizobium meliloti 1021
|
| 99 | d1jtva_ |
|
not modelled |
99.1 |
15 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 100 | d1sbya1 |
|
not modelled |
99.1 |
12 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 101 | c3iccA_ |
|
not modelled |
99.1 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative 3-oxoacyl-(acyl carrier protein) reductase;
PDBTitle: crystal structure of a putative 3-oxoacyl-(acyl carrier protein)2 reductase from bacillus anthracis at 1.87 a resolution
|
| 102 | c3lylB_ |
|
not modelled |
99.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: structure of 3-oxoacyl-acylcarrier protein reductase, fabg2 from francisella tularensis
|
| 103 | d1qsga_ |
|
not modelled |
99.1 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 104 | c2dteB_ |
|
not modelled |
99.1 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glucose 1-dehydrogenase related protein;
PDBTitle: structure of thermoplasma acidophilum aldohexose dehydrogenase (aldt)2 in complex with nadh
|
| 105 | c3un1D_ |
|
not modelled |
99.1 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:probable oxidoreductase;
PDBTitle: crystal structure of an oxidoreductase from sinorhizobium meliloti2 1021
|
| 106 | c3ijrF_ |
|
not modelled |
99.1 |
15 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family;
PDBTitle: 2.05 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor' in complex3 with nad+
|
| 107 | c3grkE_ |
|
not modelled |
99.1 |
14 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of short chain dehydrogenase reductase2 sdr glucose-ribitol dehydrogenase from brucella melitensis
|
| 108 | c3rihB_ |
|
not modelled |
99.1 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short chain dehydrogenase or reductase;
PDBTitle: crystal structure of a putative short chain dehydrogenase or reductase2 from mycobacterium abscessus
|
| 109 | c3pk0B_ |
|
not modelled |
99.0 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: crystal structure of short-chain dehydrogenase/reductase sdr from2 mycobacterium smegmatis
|
| 110 | d1uaya_ |
|
not modelled |
99.0 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 111 | c3osuA_ |
|
not modelled |
99.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of the 3-oxoacyl-acyl carrier protein reductase,2 fabg, from staphylococcus aureus
|
| 112 | c3ppiA_ |
|
not modelled |
99.0 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxyacyl-coa dehydrogenase type-2;
PDBTitle: crystal structure of 3-hydroxyacyl-coa dehydrogenase type-2 from2 mycobacterium avium
|
| 113 | c3r6dA_ |
|
not modelled |
99.0 |
15 |
PDB header:lyase, isomerase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of nad-dependent epimerase/dehydratase from2 veillonella parvula dsm 2008 with cz-methylated lysine
|
| 114 | c2uvdE_ |
|
not modelled |
99.0 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: the crystal structure of a 3-oxoacyl-(acyl carrier protein)2 reductase from bacillus anthracis (ba3989)
|
| 115 | c3k31B_ |
|
not modelled |
99.0 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of eonyl-(acyl-carrier-protein) reductase from2 anaplasma phagocytophilum in complex with nad at 1.9a resolution
|
| 116 | c3ezlA_ |
|
not modelled |
99.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:acetoacetyl-coa reductase;
PDBTitle: crystal structure of acetyacetyl-coa reductase from2 burkholderia pseudomallei 1710b
|
| 117 | c3v2gA_ |
|
not modelled |
99.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of a dehydrogenase/reductase from sinorhizobium2 meliloti 1021
|
| 118 | c3ioyB_ |
|
not modelled |
99.0 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: structure of putative short-chain dehydrogenase (saro_0793)2 from novosphingobium aromaticivorans
|
| 119 | d2ae2a_ |
|
not modelled |
99.0 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 120 | c3sx2F_ |
|
not modelled |
99.0 |
13 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative 3-ketoacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of a putative 3-ketoacyl-(acyl-carrier-protein)2 reductase from mycobacterium paratuberculosis in complex with nad
|

