1 c3giaA_
100.0
16
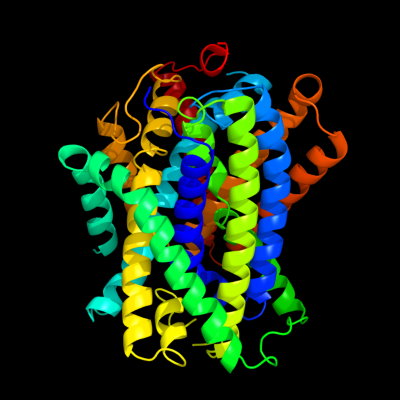
PDB header: transport proteinChain: A: PDB Molecule: uncharacterized protein mj0609;PDBTitle: crystal structure of apct transporter
2 c3lrcC_
100.0
21
PDB header: transport proteinChain: C: PDB Molecule: arginine/agmatine antiporter;PDBTitle: structure of e. coli adic (p1)
3 c2jlnA_
99.9
11
PDB header: membrane proteinChain: A: PDB Molecule: mhp1;PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
4 c2xq2A_
99.3
9
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: structure of the k294a mutant of vsglt
5 c3dh4A_
99.2
10
PDB header: transport proteinChain: A: PDB Molecule: sodium/glucose cotransporter;PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
6 d2a65a1
97.3
14
Fold: SNF-likeSuperfamily: SNF-likeFamily: SNF-like7 c2w8aC_
96.1
10
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
8 c3hfxA_
79.0
10
PDB header: transport proteinChain: A: PDB Molecule: l-carnitine/gamma-butyrobetaine antiporter;PDBTitle: crystal structure of carnitine transporter
9 c3m7bA_
57.0
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: tellurite resistance protein teha homolog;PDBTitle: crystal structure of plant slac1 homolog teha
10 c2kncA_
46.2
8
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
11 c2rddB_
40.6
26
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
12 d1fftb2
37.8
9
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region13 c3qnqD_
22.2
8
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
14 c2kluA_
21.6
14
PDB header: immune system, membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
15 c2qjkM_
20.1
10
PDB header: electron transportChain: M: PDB Molecule: cytochrome b;PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
16 c2voyG_
19.1
16
PDB header: hydrolaseChain: G: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
17 c1m57H_
18.6
12
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c oxidase;PDBTitle: structure of cytochrome c oxidase from rhodobacter2 sphaeroides (eq(i-286) mutant))
18 d1pw4a_
17.4
11
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter19 c3rkoF_
16.1
10
PDB header: oxidoreductaseChain: F: PDB Molecule: nadh-quinone oxidoreductase subunit j;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
20 c2jp3A_
15.9
11
PDB header: transcriptionChain: A: PDB Molecule: fxyd domain-containing ion transport regulator 4;PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
21 d3dtub2
not modelled
15.3
12
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region22 d3ehbb2
not modelled
15.0
19
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region23 c1qleB_
not modelled
14.6
19
PDB header: oxidoreductase/immune systemChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: cryo-structure of the paracoccus denitrificans four-subunit2 cytochrome c oxidase in the completely oxidized state3 complexed with an antibody fv fragment
24 c1ar1B_
not modelled
14.6
19
PDB header: complex (oxidoreductase/antibody)Chain: B: PDB Molecule: cytochrome c oxidase;PDBTitle: structure at 2.7 angstrom resolution of the paracoccus2 denitrificans two-subunit cytochrome c oxidase complexed3 with an antibody fv fragment
25 c1fftG_
not modelled
14.3
9
PDB header: oxidoreductaseChain: G: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
26 c1ujlA_
not modelled
13.9
10
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily hPDBTitle: solution structure of the herg k+ channel s5-p2 extracellular linker
27 c2bbjB_
not modelled
13.6
6
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
28 d3dhwa1
not modelled
13.3
12
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like29 d1jt8a_
not modelled
13.2
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like30 c3cwbQ_
not modelled
13.1
17
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
31 c3aqpB_
not modelled
13.0
11
PDB header: membrane proteinChain: B: PDB Molecule: probable secdf protein-export membrane protein;PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
32 d1iwga8
not modelled
12.5
9
Fold: Multidrug efflux transporter AcrB transmembrane domainSuperfamily: Multidrug efflux transporter AcrB transmembrane domainFamily: Multidrug efflux transporter AcrB transmembrane domain33 c3ixzA_
not modelled
11.8
12
PDB header: hydrolaseChain: A: PDB Molecule: potassium-transporting atpase alpha;PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
34 d2yvxa3
not modelled
11.8
19
Fold: MgtE membrane domain-likeSuperfamily: MgtE membrane domain-likeFamily: MgtE membrane domain-like35 d1niga_
not modelled
11.8
21
Fold: Ferritin-likeSuperfamily: Cobalamin adenosyltransferase-likeFamily: Hypothetical protein Ta123836 d1pv7a_
not modelled
11.4
10
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: LacY-like proton/sugar symporter37 c3rkoK_
not modelled
11.3
12
PDB header: oxidoreductaseChain: K: PDB Molecule: nadh-quinone oxidoreductase subunit k;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
38 c1qcrD_
not modelled
11.2
17
PDB header: PDB COMPND: 39 c2e2sA_
not modelled
11.1
11
PDB header: toxinChain: A: PDB Molecule: agelenin;PDBTitle: solution structure of agelenin, an insecticidal peptide2 from the venom of agelena opulenta
40 c2k21A_
not modelled
11.1
20
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
41 c2cpbA_
not modelled
10.8
29
PDB header: viral proteinChain: A: PDB Molecule: m13 major coat protein;PDBTitle: solution nmr structures of the major coat protein of2 filamentous bacteriophage m13 solubilized in3 dodecylphosphocholine micelles, 25 lowest energy structures
42 c2k1kB_
not modelled
10.8
17
PDB header: signaling proteinChain: B: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
43 c2k1lB_
not modelled
10.8
17
PDB header: signaling proteinChain: B: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
44 c2k1lA_
not modelled
10.8
17
PDB header: signaling proteinChain: A: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
45 c2k1kA_
not modelled
10.8
17
PDB header: signaling proteinChain: A: PDB Molecule: ephrin type-a receptor 1;PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
46 c3aa0C_
not modelled
10.0
38
PDB header: protein bindingChain: C: PDB Molecule: 21mer peptide from leucine-rich repeat-containing proteinPDBTitle: crystal structure of actin capping protein in complex with the cp-2 binding motif derived from carmil
47 c2xutC_
not modelled
9.9
7
PDB header: transport proteinChain: C: PDB Molecule: proton/peptide symporter family protein;PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
48 c2jo1A_
not modelled
9.9
11
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
49 c3b8eC_
not modelled
9.7
8
PDB header: hydrolase/transport proteinChain: C: PDB Molecule: sodium/potassium-transporting atpase subunitPDBTitle: crystal structure of the sodium-potassium pump
50 c1p84D_
not modelled
9.6
14
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
51 d1otsa_
not modelled
9.5
22
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel52 d1zela1
not modelled
9.3
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Rv2827c N-terminal domain-like53 c2jwyA_
not modelled
9.1
13
PDB header: lipoproteinChain: A: PDB Molecule: uncharacterized lipoprotein yaji;PDBTitle: solution nmr structure of uncharacterized lipoprotein yaji from2 escherichia coli. northeast structural genomics target er540
54 d2iuba2
not modelled
9.1
3
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region55 d1g2913
not modelled
9.0
29
Fold: OB-foldSuperfamily: MOP-likeFamily: ABC-transporter additional domain56 d2nr9a1
not modelled
9.0
22
Fold: Rhomboid-likeSuperfamily: Rhomboid-likeFamily: Rhomboid-like57 c3fh6F_
not modelled
8.9
13
PDB header: transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
58 c1oy8A_
not modelled
8.9
7
PDB header: membrane proteinChain: A: PDB Molecule: acriflavine resistance protein b;PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
59 c2dmpA_
not modelled
8.9
10
PDB header: dna binding proteinChain: A: PDB Molecule: zinc fingers and homeoboxes protein 2;PDBTitle: solution structure of the third homeobox domain of zinc2 fingers and homeoboxes protein 2
60 c3njcA_
not modelled
8.7
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: yslb protein;PDBTitle: crystal structure of the yslb protein from bacillus subtilis.2 northeast structural genomics consortium target sr460.
61 d1v54d_
not modelled
8.7
8
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit IVFamily: Mitochondrial cytochrome c oxidase subunit IV62 c2y69Q_
not modelled
8.6
8
PDB header: electron transportChain: Q: PDB Molecule: cytochrome c oxidase subunit 4 isoform 1;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
63 c2h3oA_
not modelled
8.5
18
PDB header: membrane proteinChain: A: PDB Molecule: merf;PDBTitle: structure of merft, a membrane protein with two trans-2 membrane helices
64 d2oara1
not modelled
8.4
17
Fold: Gated mechanosensitive channelSuperfamily: Gated mechanosensitive channelFamily: Gated mechanosensitive channel65 c2oarA_
not modelled
8.3
14
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: mechanosensitive channel of large conductance (mscl)
66 c2k1aA_
not modelled
8.2
9
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
67 c2l3hA_
not modelled
8.2
7
PDB header: hydrolaseChain: A: PDB Molecule: prostatic acid phosphatase;PDBTitle: nmr structure in a membrane environment reveals putative amyloidogenic2 regions of the sevi precursor peptide pap248-286
68 d1j9ia_
not modelled
8.2
15
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Terminase gpNU1 subunit domain69 c3ixxE_
not modelled
8.2
25
PDB header: virusChain: E: PDB Molecule: peptide pr;PDBTitle: the pseudo-atomic structure of west nile immature virus in2 complex with fab fragments of the anti-fusion loop antibody3 e53
70 c3r7gB_
not modelled
8.1
8
PDB header: protein bindingChain: B: PDB Molecule: formin-2;PDBTitle: crystal structure of spire kind domain in complex with the tail of2 fmn2
71 c2jo8B_
not modelled
8.1
27
PDB header: transferaseChain: B: PDB Molecule: serine/threonine-protein kinase 4;PDBTitle: solution structure of c-terminal domain of human mammalian2 sterile 20-like kinase 1 (mst1)
72 d1rhzb_
not modelled
8.0
18
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit73 c2cblA_
not modelled
8.0
24
PDB header: complex (proto-oncogene/peptide)Chain: A: PDB Molecule: proto-oncogene cbl;PDBTitle: n-terminal domain of cbl in complex with its binding site2 on zap-70
74 d1zh5a1
not modelled
8.0
29
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: La domain75 c3bunB_
not modelled
8.0
24
PDB header: ligase/signaling proteinChain: B: PDB Molecule: e3 ubiquitin-protein ligase cbl;PDBTitle: crystal structure of c-cbl-tkb domain complexed with its2 binding motif in sprouty4
76 d1t98a2
not modelled
7.9
21
Fold: STAT-likeSuperfamily: MukF C-terminal domain-likeFamily: MukF C-terminal domain-like77 c1wazA_
not modelled
7.8
15
PDB header: transport proteinChain: A: PDB Molecule: merf;PDBTitle: nmr structure determination of the bacterial mercury2 transporter, merf, in micelles
78 d1eysh2
not modelled
7.7
19
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region79 c3lk2T_
not modelled
7.7
50
PDB header: protein bindingChain: T: PDB Molecule: leucine-rich repeat-containing protein 16a;PDBTitle: crystal structure of capz bound to the uncapping motif from carmil
80 c3c6eC_
not modelled
7.7
43
PDB header: viral proteinChain: C: PDB Molecule: prm;PDBTitle: crystal structure of the precursor membrane protein- envelope protein2 heterodimer from the dengue 2 virus at neutral ph
81 c3mk7F_
not modelled
7.7
8
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
82 c3opyJ_
not modelled
7.6
18
PDB header: transferaseChain: J: PDB Molecule: 6-phosphofructo-1-kinase gamma-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
83 c3opyL_
not modelled
7.6
18
PDB header: transferaseChain: L: PDB Molecule: 6-phosphofructo-1-kinase gamma-subunit;PDBTitle: crystal structure of pichia pastoris phosphofructokinase in the t-2 state
84 c3p5nA_
not modelled
7.6
7
PDB header: transport proteinChain: A: PDB Molecule: riboflavin uptake protein;PDBTitle: structure and mechanism of the s component of a bacterial ecf2 transporter
85 c3hzqA_
not modelled
7.6
12
PDB header: membrane proteinChain: A: PDB Molecule: large-conductance mechanosensitive channel;PDBTitle: structure of a tetrameric mscl in an expanded intermediate2 state
86 c3c6rE_
not modelled
7.5
43
PDB header: virusChain: E: PDB Molecule: peptide pr;PDBTitle: low ph immature dengue virus
87 c2k9yB_
not modelled
7.4
13
PDB header: transferaseChain: B: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
88 c2k9yA_
not modelled
7.4
13
PDB header: transferaseChain: A: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
89 d2h8pc1
not modelled
7.3
9
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels90 c3nauA_
not modelled
7.3
11
PDB header: transcriptionChain: A: PDB Molecule: zinc fingers and homeoboxes protein 2;PDBTitle: crystal structure of zhx2 hd2 (zinc-fingers and homeoboxes protein 2,2 homeodomain 2)
91 d1zyba1
not modelled
7.2
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: CAP C-terminal domain-like92 d1rh5b_
not modelled
7.2
18
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit93 d2ecba1
not modelled
7.2
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain94 c2zxeA_
not modelled
7.2
11
PDB header: hydrolase/transport proteinChain: A: PDB Molecule: na, k-atpase alpha subunit;PDBTitle: crystal structure of the sodium - potassium pump in the e2.2k+.pi2 state
95 d2e2ca_
not modelled
7.2
14
Fold: UBC-likeSuperfamily: UBC-likeFamily: UBC-related96 c2l2tA_
not modelled
7.1
11
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-4;PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
97 c3iz5d_
not modelled
7.0
19
PDB header: ribosomeChain: D: PDB Molecule: 60s ribosomal protein l4 (l4p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
98 d1ubdc2
not modelled
7.0
43
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H299 c3op0B_
not modelled
7.0
18
PDB header: signaling protein/signaling protein reguChain: B: PDB Molecule: signal transduction protein cbl-c;PDBTitle: crystal structure of cbl-c (cbl-3) tkb domain in complex with egfr2 py1069 peptide