1 c3aehB_
100.0
18
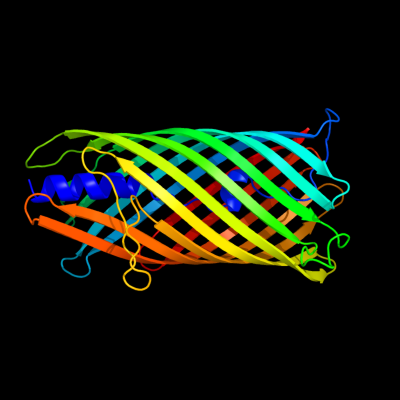
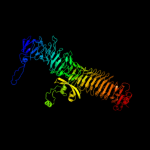
PDB header: hydrolaseChain: B: PDB Molecule: hemoglobin-binding protease hbp autotransporter;PDBTitle: integral membrane domain of autotransporter hbp
2 c3sljA_
100.0
18
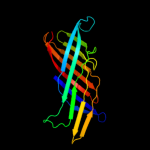
PDB header: protein transportChain: A: PDB Molecule: serine protease espp;PDBTitle: pre-cleavage structure of the autotransporter espp - n1023a mutant
3 c3qq2C_
100.0
22
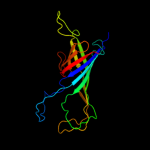
PDB header: membrane protein/protein transportChain: C: PDB Molecule: brka autotransporter;PDBTitle: crystal structure of the beta domain of the bordetella autotransporter2 brka
4 c2qomB_
100.0
18
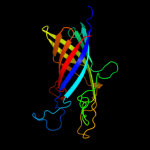
PDB header: hydrolaseChain: B: PDB Molecule: serine protease espp;PDBTitle: the crystal structure of the e.coli espp autotransporter beta-domain.
5 d1uynx_
100.0
15
Fold: Transmembrane beta-barrelsSuperfamily: AutotransporterFamily: Autotransporter6 c3kvnA_
100.0
10
PDB header: hydrolaseChain: A: PDB Molecule: esterase esta;PDBTitle: crystal structure of the full-length autotransporter esta from2 pseudomonas aeruginosa
7 d1daba_
99.9
12
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Virulence factor P.69 pertactin8 c3ml3A_
99.9
18
PDB header: protein transportChain: A: PDB Molecule: outer membrane protein icsa autotransporter;PDBTitle: crystal structure of the icsa autochaperone region
9 c3h09B_
99.7
16
PDB header: hydrolaseChain: B: PDB Molecule: immunoglobulin a1 protease;PDBTitle: the structure of haemophilus influenzae iga1 protease
10 c3syjA_
99.6
11
PDB header: cell adhesionChain: A: PDB Molecule: adhesion and penetration protein autotransporter;PDBTitle: crystal structure of the haemophilus influenzae hap adhesin
11 c3qraA_
98.3
16
PDB header: cell invasionChain: A: PDB Molecule: attachment invasion locus protein;PDBTitle: the crystal structure of ail, the attachment invasion locus protein of2 yersinia pestis
12 c2k0lA_
98.2
15
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr structure of the transmembrane domain of the outer2 membrane protein a from klebsiella pneumoniae in dhpc3 micelles.
13 d1g90a_
98.2
14
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein14 d1p4ta_
98.1
17
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein15 d1qjpa_
98.0
16
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein16 c3nb3C_
97.9
15
PDB header: virusChain: C: PDB Molecule: outer membrane protein a;PDBTitle: the host outer membrane proteins ompa and ompc are packed at specific2 sites in the shigella phage sf6 virion as structural components
17 c2jmmA_
97.7
21
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein a;PDBTitle: nmr solution structure of a minimal transmembrane beta-2 barrel platform protein
18 c2x27X_
97.4
18
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein oprg;PDBTitle: crystal structure of the outer membrane protein oprg from2 pseudomonas aeruginosa
19 d1qj8a_
96.8
10
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane protein20 c2lhfA_
96.0
23
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein h1;PDBTitle: solution structure of outer membrane protein h (oprh) from p.2 aeruginosa in dhpc micelles
21 d2pora_
not modelled
95.7
11
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin22 c2wjqA_
not modelled
95.3
11
PDB header: transport proteinChain: A: PDB Molecule: probable n-acetylneuraminic acid outer membrane channelPDBTitle: nanc porin structure in hexagonal crystal form.
23 d2zfga1
not modelled
94.9
16
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin24 c3a2rX_
not modelled
94.4
13
PDB header: membrane proteinChain: X: PDB Molecule: outer membrane protein ii;PDBTitle: crystal structure of outer membrane protein porb from neisseria2 meningitidis
25 c2iwvD_
not modelled
93.6
15
PDB header: ion channelChain: D: PDB Molecule: outer membrane protein g;PDBTitle: structure of the monomeric outer membrane porin ompg in the2 open and closed conformation
26 d3prna_
not modelled
93.1
16
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin27 d1osma_
not modelled
92.1
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin28 d2fgqx1
not modelled
90.6
13
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin29 c2f1tB_
not modelled
90.6
14
PDB header: membrane proteinChain: B: PDB Molecule: outer membrane protein w;PDBTitle: outer membrane protein ompw
30 d2vdfa1
not modelled
87.9
14
Fold: Transmembrane beta-barrelsSuperfamily: OMPT-likeFamily: Outer membrane adhesin/invasin OpcA31 c3brzA_
not modelled
83.8
12
PDB header: transport proteinChain: A: PDB Molecule: todx;PDBTitle: crystal structure of the pseudomonas putida toluene2 transporter todx
32 d1phoa_
not modelled
81.2
10
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Porin33 c3nsgA_
not modelled
68.5
11
PDB header: membrane proteinChain: A: PDB Molecule: outer membrane protein f;PDBTitle: crystal structure of ompf, an outer membrane protein from salmonella2 typhi
34 d1w0pa2
not modelled
38.4
12
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Vibrio cholerae sialidase, N-terminal and insertion domains35 c3ak5B_
not modelled
28.7
12
PDB header: hydrolaseChain: B: PDB Molecule: hemoglobin-binding protease hbp;PDBTitle: hemoglobin protease (hbp) passenger missing domain-2
36 c3rbhC_
not modelled
26.7
21
PDB header: transport proteinChain: C: PDB Molecule: alginate production protein alge;PDBTitle: structure of alginate export protein alge from pseudomonas aeruginosa
37 d1t16a_
not modelled
26.6
16
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Outer membrane protein transport protein38 d1pm6a_
not modelled
21.6
0
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Excisionase-like39 c2lfeA_
not modelled
18.0
18
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase hecw2;PDBTitle: solution nmr structure of n-terminal domain of human e3 ubiquitin-2 protein ligase hecw2, northeast structural genomics consortium (nesg)3 target ht6306a
40 c1yo8A_
not modelled
17.8
20
PDB header: cell adhesionChain: A: PDB Molecule: thrombospondin-2;PDBTitle: structure of the c-terminal domain of human thrombospondin-2
41 d1txka2
not modelled
16.1
12
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: MdoG-like42 d2mpra_
not modelled
15.7
14
Fold: Transmembrane beta-barrelsSuperfamily: PorinsFamily: Maltoporin-like43 c2x4mD_
not modelled
15.6
14
PDB header: hydrolaseChain: D: PDB Molecule: coagulase/fibrinolysin;PDBTitle: yersinia pestis plasminogen activator pla
44 c1txkA_
not modelled
13.4
12
PDB header: biosynthetic proteinChain: A: PDB Molecule: glucans biosynthesis protein g;PDBTitle: crystal structure of escherichia coli opgg
45 d1tywa_
not modelled
12.9
11
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: P22 tailspike protein46 d1vpra1
not modelled
12.3
24
Fold: LipocalinsSuperfamily: LipocalinsFamily: Dinoflagellate luciferase repeat47 d2evra2
not modelled
11.6
20
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: NlpC/P6048 c3fbyC_
not modelled
11.2
15
PDB header: cell adhesionChain: C: PDB Molecule: cartilage oligomeric matrix protein;PDBTitle: the crystal structure of the signature domain of cartilage oligomeric2 matrix protein.
49 d1upsa2
not modelled
10.9
11
Fold: beta-TrefoilSuperfamily: Ricin B-like lectinsFamily: GlcNAc-alpha-1,4-Gal-releasing endo-beta-galactosidase, GngC, C-terminal domain50 c2r5xA_
not modelled
10.7
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein;PDBTitle: crystal structure of uncharacterized conserved protein yugn from2 geobacillus kaustophilus hta426
51 c2xc1A_
not modelled
10.7
11
PDB header: hydrolaseChain: A: PDB Molecule: bifunctional tail protein;PDBTitle: full-length tailspike protein mutant y108w of bacteriophage2 p22
52 c2fg0B_
not modelled
10.6
20
PDB header: hydrolaseChain: B: PDB Molecule: cog0791: cell wall-associated hydrolases (invasion-PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
53 c1q40C_
not modelled
10.3
13
PDB header: translationChain: C: PDB Molecule: mrna transport regulator mtr2;PDBTitle: crystal structure of the c. albicans mtr2-mex67 m domain complex
54 d1rh6a_
not modelled
9.5
0
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: Excisionase-like55 d2pwwa1
not modelled
9.4
20
Fold: TBP-likeSuperfamily: YugN-likeFamily: YugN-like56 d3ejva1
not modelled
9.4
23
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: BaiE/LinA-like57 c2bvbA_
not modelled
9.1
19
PDB header: adhesionChain: A: PDB Molecule: micronemal protein 1;PDBTitle: the c-terminal domain from micronemal protein 1 (mic1) from2 toxoplasma gondii
58 c3emoA_
not modelled
9.0
16
PDB header: membrane protein/cell adhesionChain: A: PDB Molecule: hia (adhesin);PDBTitle: crystal structure of transmembrane hia 973-1098
59 c1lj2B_
not modelled
8.8
45
PDB header: viral protein/ translationChain: B: PDB Molecule: nonstructural rna-binding protein 34;PDBTitle: recognition of eif4g by rotavirus nsp3 reveals a basis for2 mrna circularization
60 c3dwoX_
not modelled
8.7
12
PDB header: membrane proteinChain: X: PDB Molecule: probable outer membrane protein;PDBTitle: crystal structure of a pseudomonas aeruginosa fadl homologue
61 c1po3A_
not modelled
8.2
11
PDB header: membrane proteinChain: A: PDB Molecule: iron(iii) dicitrate transport protein fecaPDBTitle: crystal structure of ferric citrate transporter feca in2 complex with ferric citrate
62 c2k4tA_
not modelled
7.8
13
PDB header: membrane protein,apoptosisChain: A: PDB Molecule: voltage-dependent anion-selective channelPDBTitle: solution structure of human vdac-1 in ldao micelles
63 c2gr7C_
not modelled
7.4
16
PDB header: membrane proteinChain: C: PDB Molecule: adhesin;PDBTitle: hia 992-1098
64 d2gr7a1
not modelled
7.4
16
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: YadA C-terminal domain-like65 c2npiD_
not modelled
7.1
57
PDB header: transcriptionChain: D: PDB Molecule: protein pcf11;PDBTitle: clp1-atp-pcf11 complex
66 d1zrua3
not modelled
6.7
50
Fold: Pseudo beta-prismSuperfamily: Bacteriophage trimeric proteins domainFamily: Lactophage receptor-binding protein N-terminal domain67 c2v5iA_
not modelled
6.4
8
PDB header: viral proteinChain: A: PDB Molecule: salmonella typhimurium db7155 bacteriophage det7PDBTitle: structure of the receptor-binding protein of bacteriophage2 det7: a podoviral tailspike in a myovirus
68 d1vjpa2
not modelled
6.3
13
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like69 c2d4nA_
not modelled
6.2
15
PDB header: hydrolaseChain: A: PDB Molecule: du;PDBTitle: crystal structure of m-pmv dutpase complexed with dupnpp, substrate2 analogue
70 d2gr8a1
not modelled
6.2
16
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: YadA C-terminal domain-like71 d2h1ta1
not modelled
6.0
19
Fold: Spiral beta-rollSuperfamily: PA1994-likeFamily: PA1994-like72 c2fhdA_
not modelled
6.0
23
PDB header: cell cycleChain: A: PDB Molecule: dna repair protein rhp9/crb2;PDBTitle: crystal structure of crb2 tandem tudor domains
73 c3cinA_
not modelled
6.0
12
PDB header: isomeraseChain: A: PDB Molecule: myo-inositol-1-phosphate synthase-related protein;PDBTitle: crystal structure of a myo-inositol-1-phosphate synthase-related2 protein (tm_1419) from thermotoga maritima msb8 at 1.70 a resolution
74 c2k1gA_
not modelled
5.9
35
PDB header: lipoproteinChain: A: PDB Molecule: lipoprotein spr;PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
75 d1jmxa4
not modelled
5.9
39
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Quinohemoprotein amine dehydrogenase A chain, domains 4 and 576 d1p1ja2
not modelled
5.9
25
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Dihydrodipicolinate reductase-like77 d2h7za1
not modelled
5.9
29
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins78 d1k1za_
not modelled
5.8
11
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain79 d1zvca1
not modelled
5.8
15
Fold: AOC barrel-likeSuperfamily: Allene oxide cyclase-likeFamily: Allene oxide cyclase-like80 c2odlA_
not modelled
5.8
11
PDB header: cell adhesionChain: A: PDB Molecule: adhesin;PDBTitle: crystal structure of the hmw1 secretion domain from2 haemophilus influenzae
81 c1w0pA_
not modelled
5.7
18
PDB header: hydrolaseChain: A: PDB Molecule: sialidase;PDBTitle: vibrio cholerae sialidase with alpha-2,6-sialyllactose
82 c2o18A_
not modelled
5.7
16
PDB header: lipid binding proteinChain: A: PDB Molecule: thiamine biosynthesis lipoprotein apbe;PDBTitle: crystal structure of a thiamine biosynthesis lipoprotein2 apbe, northeast strcutural genomics target er559
83 c1dfcB_
not modelled
5.6
11
PDB header: structural proteinChain: B: PDB Molecule: fascin;PDBTitle: crystal structure of human fascin, an actin-crosslinking protein
84 d1gcqc_
not modelled
5.6
10
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain85 d1ylea1
not modelled
5.6
9
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: AstA-like86 d2g03a1
not modelled
5.6
14
Fold: Fic-likeSuperfamily: Fic-likeFamily: Fic-like87 d1sm3h2
not modelled
5.6
4
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: C1 set domains (antibody constant domain-like)88 c1xkwA_
not modelled
5.4
10
PDB header: membrane proteinChain: A: PDB Molecule: fe(iii)-pyochelin receptor;PDBTitle: pyochelin outer membrane receptor fpta from pseudomonas2 aeruginosa
89 d2ihoa2
not modelled
5.3
17
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: MOA C-terminal domain-like90 c3n0kA_
not modelled
5.3
13
PDB header: hydrolase inhibitorChain: A: PDB Molecule: serine protease inhibitor 1;PDBTitle: proteinase inhibitor from coprinopsis cinerea