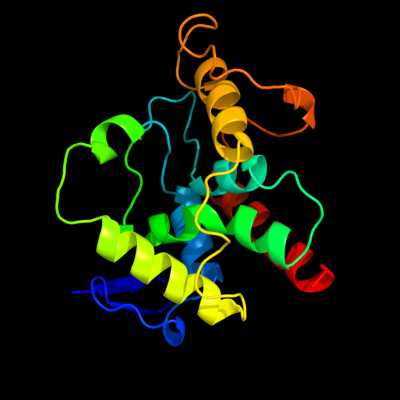
| 1 | d2gz4a1
|
|
 |
100.0 |
29 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
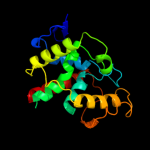
| 2 | c3mzoA_
|
|
 |
99.4 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:lin2634 protein;
PDBTitle: crystal structure of a hd-domain phosphohydrolase (lin2634) from2 listeria innocua at 1.98 a resolution
|
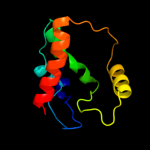
| 3 | d2paqa1
|
|
 |
99.2 |
18 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
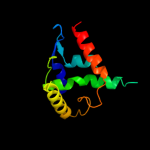
| 4 | c2cqzA_
|
|
 |
98.8 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:177aa long hypothetical protein;
PDBTitle: crystal structure of ph0347 protein from pyrococcus horikoshii ot3
|
| 5 | d1xx7a_
|
|
 |
98.8 |
25 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 6 | d1ynba1
|
|
 |
98.2 |
21 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 7 | c3kh1B_
|
|
 |
98.1 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:predicted metal-dependent phosphohydrolase;
PDBTitle: crystal structure of predicted metal-dependent2 phosphohydrolase (zp_00055740.2) from magnetospirillum3 magnetotacticum ms-1 at 1.37 a resolution
|
| 8 | d3b57a1
|
|
 |
80.3 |
17 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 9 | c2ogiA_
|
|
 |
76.2 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein sag1661;
PDBTitle: crystal structure of a putative metal dependent phosphohydrolase2 (sag1661) from streptococcus agalactiae serogroup v at 1.85 a3 resolution
|
| 10 | c2pgsA_
|
|
 |
73.1 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative deoxyguanosinetriphosphate triphosphohydrolase;
PDBTitle: crystal structure of a putative deoxyguanosinetriphosphate2 triphosphohydrolase from pseudomonas syringae pv. phaseolicola 1448a
|
| 11 | c2o08B_
|
|
 |
71.2 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:bh1327 protein;
PDBTitle: crystal structure of a putative hd superfamily hydrolase (bh1327) from2 bacillus halodurans at 1.90 a resolution
|
| 12 | c3ccgA_
|
|
 |
70.6 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:hd superfamily hydrolase;
PDBTitle: crystal structure of predicted hd superfamily hydrolase involved in2 nad metabolism (np_347894.1) from clostridium acetobutylicum at 1.503 a resolution
|
| 13 | d2pq7a1
|
|
 |
70.2 |
17 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 14 | c3nr1A_
|
|
 |
65.8 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:hd domain-containing protein 3;
PDBTitle: a metazoan ortholog of spot hydrolyzes ppgpp and plays a role in2 starvation responses
|
| 15 | c1vj7B_
|
|
 |
58.9 |
13 |
PDB header:hydrolase, transferase
Chain: B: PDB Molecule:bifunctional rela/spot;
PDBTitle: crystal structure of the bifunctional catalytic fragment of relseq,2 the rela/spot homolog from streptococcus equisimilis.
|
| 16 | c2dqbB_
|
|
 |
49.5 |
36 |
PDB header:hydrolase, dna binding protein
Chain: B: PDB Molecule:deoxyguanosinetriphosphate triphosphohydrolase, putative;
PDBTitle: crystal structure of dntp triphosphohydrolase from thermus2 thermophilus hb8, which is homologous to dgtp triphosphohydrolase
|
| 17 | c3m1tA_
|
|
 |
48.6 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative phosphohydrolase;
PDBTitle: crystal structure of putative phosphohydrolase (yp_929327.1) from2 shewanella amazonensis sb2b at 1.62 a resolution
|
| 18 | d2heka1
|
|
 |
47.5 |
23 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 19 | d2pjqa1
|
|
 |
45.6 |
25 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 20 | c3nqwB_
|
|
 |
42.4 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:cg11900;
PDBTitle: a metazoan ortholog of spot hydrolyzes ppgpp and plays a role in2 starvation responses
|
| 21 | c3hc1A_ |
|
not modelled |
41.8 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized hdod domain protein;
PDBTitle: crystal structure of hdod domain protein with unknown function2 (np_953345.1) from geobacter sulfurreducens at 1.90 a resolution
|
| 22 | c3u1nC_ |
|
not modelled |
40.2 |
36 |
PDB header:hydrolase
Chain: C: PDB Molecule:sam domain and hd domain-containing protein 1;
PDBTitle: structure of the catalytic core of human samhd1
|
| 23 | d3dtoa1 |
|
not modelled |
38.3 |
19 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 24 | d1vqra_ |
|
not modelled |
36.0 |
11 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:modified HD domain |
| 25 | d2qgsa1 |
|
not modelled |
29.3 |
18 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 26 | d1rm6c1 |
|
not modelled |
29.0 |
29 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
| 27 | c2o6iA_ |
|
not modelled |
28.5 |
41 |
PDB header:hydrolase
Chain: A: PDB Molecule:hd domain protein;
PDBTitle: structure of an enterococcus faecalis hd domain phosphohydrolase
|
| 28 | d2o6ia1 |
|
not modelled |
28.5 |
41 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 29 | c3aqnA_ |
|
not modelled |
27.5 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:poly(a) polymerase;
PDBTitle: complex structure of bacterial protein (apo form ii)
|
| 30 | d1n62a1 |
|
not modelled |
27.3 |
24 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
| 31 | c2q14A_ |
|
not modelled |
27.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphohydrolase;
PDBTitle: crystal structure of phosphohydrolase (bt4208) from bacteroides2 thetaiotaomicron vpi-5482 at 2.20 a resolution
|
| 32 | d1zxia1 |
|
not modelled |
26.3 |
24 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
| 33 | c3hrdH_ |
|
not modelled |
22.3 |
38 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:nicotinate dehydrogenase small fes subunit;
PDBTitle: crystal structure of nicotinate dehydrogenase
|
| 34 | d3djba1 |
|
not modelled |
21.1 |
30 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 35 | d1ffva1 |
|
not modelled |
20.6 |
29 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
| 36 | c1ffuA_ |
|
not modelled |
20.4 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:cuts, iron-sulfur protein of carbon monoxide
PDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava which lacks the mo-pyranopterin moiety of the3 molybdenum cofactor
|
| 37 | c3memA_ |
|
not modelled |
20.2 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:putative signal transduction protein;
PDBTitle: crystal structure of a putative signal transduction protein2 (maqu_0641) from marinobacter aquaeolei vt8 at 2.25 a resolution
|
| 38 | d1t3qa1 |
|
not modelled |
19.5 |
24 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
| 39 | c3l09B_ |
|
not modelled |
19.5 |
11 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of putative transcriptional regulator2 (jann_22dec04_contig27_revised_gene3569) from jannaschia sp. ccs1 at3 2.81 a resolution
|
| 40 | c3gw7A_ |
|
not modelled |
17.4 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein yedj;
PDBTitle: crystal structure of a metal-dependent phosphohydrolase2 with conserved hd domain (yedj) from escherichia coli in3 complex with nickel ions. northeast structural genomics4 consortium target er63
|
| 41 | c3skdA_ |
|
not modelled |
17.2 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein tthb187;
PDBTitle: crystal structure of the thermus thermophilus cas3 hd domain in the2 presence of ni2+
|
| 42 | c1t3qD_ |
|
not modelled |
17.2 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:quinoline 2-oxidoreductase small subunit;
PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
|
| 43 | d1v97a1 |
|
not modelled |
17.0 |
29 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
| 44 | d1b93a_ |
|
not modelled |
17.0 |
11 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 45 | c3bg2A_ |
|
not modelled |
16.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:dgtp triphosphohydrolase;
PDBTitle: crystal structure of deoxyguanosinetriphosphate triphosphohydrolase2 from flavobacterium sp. med217
|
| 46 | d1vj7a1 |
|
not modelled |
16.4 |
13 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 47 | d1vmda_ |
|
not modelled |
15.8 |
16 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 48 | c1n60D_ |
|
not modelled |
13.6 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:carbon monoxide dehydrogenase small chain;
PDBTitle: crystal structure of the cu,mo-co dehydrogenase (codh); cyanide-2 inactivated form
|
| 49 | c2jo7A_ |
|
not modelled |
12.9 |
23 |
PDB header:surface active protein
Chain: A: PDB Molecule:glycosylphosphatidylinositol-anchored merozoite
PDBTitle: solution structure of the adhesion protein bd37 from2 babesia divergens
|
| 50 | c1rm6F_ |
|
not modelled |
11.7 |
29 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:4-hydroxybenzoyl-coa reductase gamma subunit;
PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
|
| 51 | d1dgja1 |
|
not modelled |
11.5 |
25 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
| 52 | c3ljvA_ |
|
not modelled |
11.4 |
18 |
PDB header:transcription
Chain: A: PDB Molecule:mmoq response regulator;
PDBTitle: crystal structure of mmoq response regulator (fragment 29-302) from2 methylococcus capsulatus str. bath, northeast structural genomics3 consortium target mcr175m
|
| 53 | d1z2cb1 |
|
not modelled |
10.7 |
17 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Diap1 N-terninal region-like |
| 54 | d1vlba1 |
|
not modelled |
10.3 |
21 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
| 55 | d1ei1a1 |
|
not modelled |
10.3 |
11 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:DNA gyrase/MutL, second domain |
| 56 | d1tuza_ |
|
not modelled |
9.2 |
18 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:EF-hand modules in multidomain proteins |
| 57 | c2zklA_ |
|
not modelled |
9.1 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:capsular polysaccharide synthesis enzyme cap5f;
PDBTitle: crystal structure of capsular polysaccharide assembling protein capf2 from staphylococcus aureus
|
| 58 | c3eu8D_ |
|
not modelled |
8.9 |
20 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative glucoamylase;
PDBTitle: crystal structure of putative glucoamylase (yp_210071.1) from2 bacteroides fragilis nctc 9343 at 2.12 a resolution
|
| 59 | c2lbbA_ |
|
not modelled |
8.5 |
11 |
PDB header:protein binding
Chain: A: PDB Molecule:acyl coa binding protein;
PDBTitle: solution structure of acyl coa binding protein from babesia bovis t2bo
|
| 60 | c2bnxA_ |
|
not modelled |
8.4 |
17 |
PDB header:structural protein
Chain: A: PDB Molecule:diaphanous protein homolog 1;
PDBTitle: crystal structure of the dimeric regulatory domain of mouse2 diaphaneous-related formin (drf), mdia1
|
| 61 | d2bnxa1 |
|
not modelled |
8.4 |
17 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:Diap1 N-terninal region-like |
| 62 | d2enda_ |
|
not modelled |
8.0 |
50 |
Fold:T4 endonuclease V
Superfamily:T4 endonuclease V
Family:T4 endonuclease V |
| 63 | c1vlbA_ |
|
not modelled |
7.8 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldehyde oxidoreductase;
PDBTitle: structure refinement of the aldehyde oxidoreductase from2 desulfovibrio gigas at 1.28 a
|
| 64 | d1kija1 |
|
not modelled |
7.7 |
12 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:DNA gyrase/MutL, second domain |
| 65 | d1mx3a2 |
|
not modelled |
7.7 |
33 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
| 66 | d1b4ua_ |
|
not modelled |
7.6 |
11 |
Fold:LigA subunit of an aromatic-ring-opening dioxygenase LigAB
Superfamily:LigA subunit of an aromatic-ring-opening dioxygenase LigAB
Family:LigA subunit of an aromatic-ring-opening dioxygenase LigAB |
| 67 | c2it8A_ |
|
not modelled |
7.5 |
67 |
PDB header:plant protein
Chain: A: PDB Molecule:trypsin inhibitor 2;
PDBTitle: solution structure of a linear analog of the cyclic squash2 trypsin inhibitor mcoti-ii
|
| 68 | d2it8a1 |
|
not modelled |
7.5 |
67 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Plant inhibitors of proteinases and amylases
Family:Plant inhibitors of proteinases and amylases |
| 69 | c2copA_ |
|
not modelled |
7.4 |
16 |
PDB header:lipid binding protein
Chain: A: PDB Molecule:acyl-coenzyme a binding domain containing 6;
PDBTitle: solution structure of rsgi ruh-040, an acbp domain from2 human cdna
|
| 70 | d2ibna1 |
|
not modelled |
7.4 |
27 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:MioX-like |
| 71 | c2huoA_ |
|
not modelled |
7.3 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inositol oxygenase;
PDBTitle: crystal structure of mouse myo-inositol oxygenase in complex with2 substrate
|
| 72 | d3bxda1 |
|
not modelled |
7.3 |
24 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:MioX-like |
| 73 | d1ivza_ |
|
not modelled |
7.0 |
5 |
Fold:Ferredoxin-like
Superfamily:SEA domain
Family:SEA domain |
| 74 | d1y8xb1 |
|
not modelled |
6.8 |
31 |
Fold:Activating enzymes of the ubiquitin-like proteins
Superfamily:Activating enzymes of the ubiquitin-like proteins
Family:Ubiquitin activating enzymes (UBA) |
| 75 | c3i7aA_ |
|
not modelled |
6.8 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative metal-dependent phosphohydrolase;
PDBTitle: crystal structure of putative metal-dependent phosphohydrolase2 (yp_926882.1) from shewanella amazonensis sb2b at 2.06 a resolution
|
| 76 | c3fnvB_ |
|
not modelled |
6.8 |
19 |
PDB header:metal binding protein
Chain: B: PDB Molecule:cdgsh iron sulfur domain-containing protein 2;
PDBTitle: crystal structure of miner1: the redox-active 2fe-2s protein causative2 in wolfram syndrome 2
|
| 77 | c3ojlA_ |
|
not modelled |
6.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cap5o;
PDBTitle: native structure of the udp-n-acetyl-mannosamine dehydrogenase cap5o2 from staphylococcus aureus
|
| 78 | c4a2iV_ |
|
not modelled |
6.7 |
11 |
PDB header:ribosome/hydrolase
Chain: V: PDB Molecule:putative ribosome biogenesis gtpase rsga;
PDBTitle: cryo-electron microscopy structure of the 30s subunit in complex with2 the yjeq biogenesis factor
|
| 79 | d1ha9a_ |
|
not modelled |
6.5 |
67 |
Fold:Knottins (small inhibitors, toxins, lectins)
Superfamily:Plant inhibitors of proteinases and amylases
Family:Plant inhibitors of proteinases and amylases |
| 80 | d1hbka_ |
|
not modelled |
6.3 |
11 |
Fold:Acyl-CoA binding protein-like
Superfamily:Acyl-CoA binding protein
Family:Acyl-CoA binding protein |
| 81 | d1dw0a_ |
|
not modelled |
6.3 |
21 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:monodomain cytochrome c |
| 82 | c2cquA_ |
|
not modelled |
6.2 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:peroxisomal d3,d2-enoyl-coa isomerase;
PDBTitle: solution structure of rsgi ruh-045, a human acyl-coa2 binding protein
|
| 83 | c3gwnA_ |
|
not modelled |
6.1 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable fad-linked sulfhydryl oxidase r596;
PDBTitle: crystal structure of the fad binding domain from mimivirus sulfhydryl2 oxidase r596
|
| 84 | d2raqa1 |
|
not modelled |
6.1 |
23 |
Fold:Ferredoxin-like
Superfamily:MTH889-like
Family:MTH889-like |
| 85 | c2po8A_ |
|
not modelled |
6.1 |
67 |
PDB header:plant protein
Chain: A: PDB Molecule:mcoti-ii;
PDBTitle: the structure of a two-disulfide intermediate of mcoti-ii
|
| 86 | d1hjra_ |
|
not modelled |
6.0 |
16 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:RuvC resolvase |
| 87 | c1tg6G_ |
|
not modelled |
5.9 |
8 |
PDB header:hydrolase
Chain: G: PDB Molecule:putative atp-dependent clp protease proteolytic subunit;
PDBTitle: crystallography and mutagenesis point to an essential role for the n-2 terminus of human mitochondrial clpp
|
| 88 | c1m3wA_ |
|
not modelled |
5.8 |
30 |
PDB header:de novo protein
Chain: A: PDB Molecule:h10h24;
PDBTitle: crystal structure of a molecular maquette scaffold
|
| 89 | c1m3wB_ |
|
not modelled |
5.8 |
30 |
PDB header:de novo protein
Chain: B: PDB Molecule:h10h24;
PDBTitle: crystal structure of a molecular maquette scaffold
|
| 90 | c1m3wC_ |
|
not modelled |
5.8 |
30 |
PDB header:de novo protein
Chain: C: PDB Molecule:h10h24;
PDBTitle: crystal structure of a molecular maquette scaffold
|
| 91 | c1m3wD_ |
|
not modelled |
5.8 |
30 |
PDB header:de novo protein
Chain: D: PDB Molecule:h10h24;
PDBTitle: crystal structure of a molecular maquette scaffold
|
| 92 | d1hyoa1 |
|
not modelled |
5.7 |
29 |
Fold:SH3-like barrel
Superfamily:Fumarylacetoacetate hydrolase, FAH, N-terminal domain
Family:Fumarylacetoacetate hydrolase, FAH, N-terminal domain |
| 93 | c3kfwX_ |
|
not modelled |
5.7 |
12 |
PDB header:structural genomics, unknown function
Chain: X: PDB Molecule:uncharacterized protein;
PDBTitle: uncharacterized protein rv0674 from mycobacterium tuberculosis
|
| 94 | c1dgjA_ |
|
not modelled |
5.5 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldehyde oxidoreductase;
PDBTitle: crystal structure of the aldehyde oxidoreductase from2 desulfovibrio desulfuricans atcc 27774
|
| 95 | d1jroa1 |
|
not modelled |
5.4 |
40 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
| 96 | c3q7hM_ |
|
not modelled |
5.4 |
13 |
PDB header:hydrolase
Chain: M: PDB Molecule:atp-dependent clp protease proteolytic subunit;
PDBTitle: structure of the clpp subunit of the atp-dependent clp protease from2 coxiella burnetii
|
| 97 | d1tg6a1 |
|
not modelled |
5.3 |
8 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Clp protease, ClpP subunit |
| 98 | d1hb6a_ |
|
not modelled |
5.2 |
11 |
Fold:Acyl-CoA binding protein-like
Superfamily:Acyl-CoA binding protein
Family:Acyl-CoA binding protein |