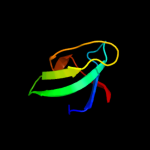
1 c2vv5D_
100.0
20
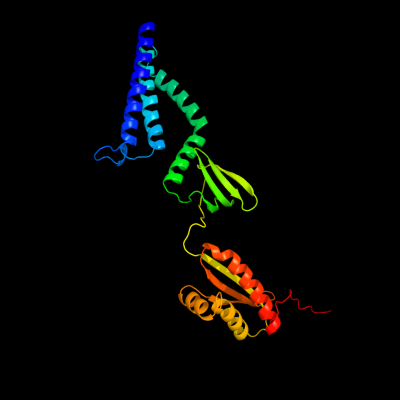
PDB header: membrane proteinChain: D: PDB Molecule: small-conductance mechanosensitive channel;PDBTitle: the open structure of mscs
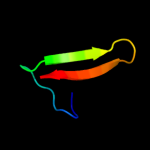
2 d2vv5a2
99.4
16
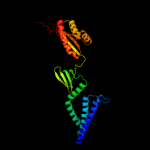
Fold: Ferredoxin-likeSuperfamily: Mechanosensitive channel protein MscS (YggB), C-terminal domainFamily: Mechanosensitive channel protein MscS (YggB), C-terminal domain3 d2vv5a1
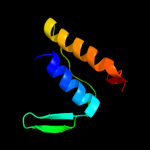
99.0
28
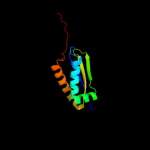
Fold: Sm-like foldSuperfamily: Sm-like ribonucleoproteinsFamily: Mechanosensitive channel protein MscS (YggB), middle domain4 d2vv5a3
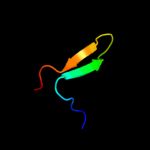
98.2
19
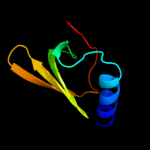
Fold: Mechanosensitive channel protein MscS (YggB), transmembrane regionSuperfamily: Mechanosensitive channel protein MscS (YggB), transmembrane regionFamily: Mechanosensitive channel protein MscS (YggB), transmembrane region5 c2e6zA_
78.1
16
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
6 d1nz9a_
77.8
33
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain7 c2vfyA_
77.5
13
PDB header: hydrolaseChain: A: PDB Molecule: akap18 delta;PDBTitle: akap18 delta central domain
8 c2kvqG_
76.9
22
PDB header: transcriptionChain: G: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of nuse:nusg-ctd complex
9 c2jvvA_
76.9
22
PDB header: transcriptionChain: A: PDB Molecule: transcription antitermination protein nusg;PDBTitle: solution structure of e. coli nusg carboxyterminal domain
10 d1nppa2
76.4
43
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: N-utilization substance G protein NusG, C-terminal domain11 d2hqha1
75.6
29
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain12 d2dfaa1
75.2
23
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: LamB/YcsF-like13 c2zkrt_
71.8
16
PDB header: ribosomal protein/rnaChain: T: PDB Molecule: rna expansion segment es39 part iii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
14 c3p8bB_
69.9
20
PDB header: transferase/transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: x-ray crystal structure of pyrococcus furiosus transcription2 elongation factor spt4/5
15 d2cqaa1
69.0
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TIP49 domain16 d1v6ta_
68.2
18
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: LamB/YcsF-like17 d2cp6a1
66.4
30
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain18 c1oy8A_
65.8
13
PDB header: membrane proteinChain: A: PDB Molecule: acriflavine resistance protein b;PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
19 c2d5wA_
65.5
15
PDB header: peptide binding proteinChain: A: PDB Molecule: peptide abc transporter, peptide-binding protein;PDBTitle: the crystal structure of oligopeptide binding protein from thermus2 thermophilus hb8 complexed with pentapeptide
20 d1t9ha1
64.3
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like21 d1u0la1
not modelled
63.0
25
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like22 c2x5cB_
not modelled
62.8
21
PDB header: viral proteinChain: B: PDB Molecule: hypothetical protein orf131;PDBTitle: crystal structure of hypothetical protein orf131 from2 pyrobaculum spherical virus
23 c3ftjA_
not modelled
60.3
18
PDB header: hydrolaseChain: A: PDB Molecule: macrolide export atp-binding/permease proteinPDBTitle: crystal structure of the periplasmic region of macb from2 actinobacillus actinomycetemcomitans
24 d1whka_
not modelled
59.7
17
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain25 c3ry3B_
not modelled
58.1
20
PDB header: transport proteinChain: B: PDB Molecule: putative solute-binding protein;PDBTitle: putative solute-binding protein from yersinia pestis.
26 d1vqot1
not modelled
55.9
30
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e27 d1h6za2
not modelled
55.0
26
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain28 c3pamB_
not modelled
54.5
18
PDB header: transport proteinChain: B: PDB Molecule: transmembrane protein;PDBTitle: crystal structure of a domain of transmembrane protein of abc-type2 oligopeptide transport system from bartonella henselae str. houston-1
29 c2rcnA_
not modelled
54.3
31
PDB header: hydrolaseChain: A: PDB Molecule: probable gtpase engc;PDBTitle: crystal structure of the ribosomal interacting gtpase yjeq from the2 enterobacterial species salmonella typhimurium.
30 c4a1cS_
not modelled
53.1
25
PDB header: ribosomeChain: S: PDB Molecule: rpl26;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
31 d1vqoq1
not modelled
52.7
30
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e32 d2coya1
not modelled
51.6
23
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain33 d1whma_
not modelled
51.5
18
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain34 d2p13a1
not modelled
51.0
13
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like35 d1whja_
not modelled
50.5
33
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain36 c1t9hA_
not modelled
50.4
21
PDB header: hydrolaseChain: A: PDB Molecule: probable gtpase engc;PDBTitle: the crystal structure of yloq, a circularly permuted gtpase.
37 d2do3a1
not modelled
50.2
19
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: SPT5 KOW domain-like38 d2e3ia1
not modelled
50.2
15
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain39 d2cp5a1
not modelled
50.1
14
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain40 d2cp2a1
not modelled
48.8
25
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain41 c2e4hA_
not modelled
48.8
27
PDB header: structural proteinChain: A: PDB Molecule: restin;PDBTitle: solution structure of cytoskeletal protein in complex with2 tubulin tail
42 d2plsa1
not modelled
48.6
20
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like43 c1m1gB_
not modelled
48.3
43
PDB header: transcriptionChain: B: PDB Molecule: transcription antitermination protein nusg;PDBTitle: crystal structure of aquifex aeolicus n-utilization2 substance g (nusg), space group p2(1)
44 c2z0wA_
not modelled
48.1
20
PDB header: protein bindingChain: A: PDB Molecule: cap-gly domain-containing linker protein 4;PDBTitle: crystal structure of the 2nd cap-gly domain in human restin-2 like protein 2 reveals a swapped-dimer
45 d1khia1
not modelled
47.1
23
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: eIF5a N-terminal domain-like46 d2o1ra1
not modelled
47.0
14
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like47 d2coza1
not modelled
47.0
27
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain48 d1bkba1
not modelled
46.9
21
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: eIF5a N-terminal domain-like49 d2e3ha1
not modelled
46.9
30
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain50 d1ppje2
not modelled
46.7
4
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor51 c3nkuA_
not modelled
45.9
28
PDB header: protein transportChain: A: PDB Molecule: drra;PDBTitle: crystal structure of the n-terminal domain of drra/sidm from2 legionella pneumophila
52 c2omdB_
not modelled
45.7
13
PDB header: lyaseChain: B: PDB Molecule: molybdopterin-converting factor subunit 2;PDBTitle: crystal structure of molybdopterin converting factor subunit 22 (aq_2181) from aquifex aeolicus vf5
53 c2jxfA_
not modelled
45.1
18
PDB header: viral protein, membrane proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: the solution structure of hcv ns4b(40-69)
54 c3iz5Y_
not modelled
45.1
19
PDB header: ribosomeChain: Y: PDB Molecule: 60s ribosomal protein l26 (l24p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
55 c3ftoA_
not modelled
45.1
16
PDB header: peptide binding proteinChain: A: PDB Molecule: oligopeptide-binding protein oppa;PDBTitle: crystal structure of oppa in a open conformation
56 d1dpea_
not modelled
44.9
25
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like57 c2qieA_
not modelled
44.8
16
PDB header: transferaseChain: A: PDB Molecule: molybdopterin-converting factor subunit 2;PDBTitle: staphylococcus aureus molybdopterin synthase in complex2 with precursor z
58 c2grvC_
not modelled
44.6
17
PDB header: biosynthetic proteinChain: C: PDB Molecule: lpqw;PDBTitle: crystal structure of lpqw
59 d1kbla2
not modelled
44.5
19
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain60 d1iuha_
not modelled
44.3
12
Fold: LigT-likeSuperfamily: LigT-likeFamily: 2'-5' RNA ligase LigT61 c3tpaA_
not modelled
44.2
14
PDB header: heme binding proteinChain: A: PDB Molecule: heme-binding protein a;PDBTitle: structure of hbpa2 from haemophilus parasuis
62 d1iz6a1
not modelled
43.4
17
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: eIF5a N-terminal domain-like63 d1u7ka_
not modelled
43.2
20
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain64 d1s04a_
not modelled
43.2
18
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain65 d2cowa1
not modelled
43.1
32
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain66 c1u0lB_
not modelled
42.8
25
PDB header: hydrolaseChain: B: PDB Molecule: probable gtpase engc;PDBTitle: crystal structure of yjeq from thermotoga maritima
67 d1iwga1
not modelled
42.4
16
Fold: Ferredoxin-likeSuperfamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomainsFamily: Multidrug efflux transporter AcrB pore domain; PN1, PN2, PC1 and PC2 subdomains68 d1xoca1
not modelled
42.3
20
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like69 d2cp0a1
not modelled
42.0
31
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain70 c2wctC_
not modelled
41.9
32
PDB header: rna-binding proteinChain: C: PDB Molecule: non-structural protein 3;PDBTitle: human sars coronavirus unique domain (triclinic form)
71 d1vbga2
not modelled
41.0
19
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain72 c3dedB_
not modelled
40.9
13
PDB header: membrane proteinChain: B: PDB Molecule: probable hemolysin;PDBTitle: c-terminal domain of probable hemolysin from chromobacterium violaceum
73 d1vr5a1
not modelled
40.9
12
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like74 c3lvuB_
not modelled
40.4
17
PDB header: transport proteinChain: B: PDB Molecule: abc transporter, periplasmic substrate-binding protein;PDBTitle: crystal structure of abc transporter, periplasmic substrate-binding2 protein spo2066 from silicibacter pomeroyi
75 c3iz5U_
not modelled
40.3
34
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l21 (l21e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
76 c2o7jA_
not modelled
40.0
18
PDB header: sugar binding proteinChain: A: PDB Molecule: oligopeptide abc transporter, periplasmicPDBTitle: the x-ray crystal structure of a thermophilic cellobiose2 binding protein bound with cellopentaose
77 c4a1aP_
not modelled
39.6
41
PDB header: ribosomeChain: P: PDB Molecule: 60s ribosomal protein l21;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
78 d2cp3a1
not modelled
39.6
35
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain79 d2eyqa1
not modelled
39.6
16
Fold: SH3-like barrelSuperfamily: CarD-likeFamily: CarD-like80 d3deda1
not modelled
39.6
13
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like81 d1whha_
not modelled
39.3
22
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain82 d1xnea_
not modelled
39.2
15
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain83 c2e70A_
not modelled
39.2
20
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the fifth kow motif of human2 transcription elongation factor spt5
84 c1ztyA_
not modelled
39.0
14
PDB header: sugar binding protein, signaling proteinChain: A: PDB Molecule: chitin oligosaccharide binding protein;PDBTitle: crystal structure of the chitin oligasaccharide binding2 protein
85 d2nqwa1
not modelled
39.0
19
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like86 d1m9dc_
not modelled
38.9
21
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain87 d2eifa1
not modelled
38.7
17
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: eIF5a N-terminal domain-like88 d1v6ga2
not modelled
37.9
27
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain89 d1yvca1
not modelled
37.7
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TRAM domain90 d1e32a3
not modelled
37.7
18
Fold: Cdc48 domain 2-likeSuperfamily: Cdc48 domain 2-likeFamily: Cdc48 domain 2-like91 c3llbA_
not modelled
37.2
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of the protein pa3983 with unknown2 function from pseudomonas aeruginosa pao1
92 d1m9fd_
not modelled
37.0
15
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain93 c2yv5A_
not modelled
36.6
16
PDB header: hydrolaseChain: A: PDB Molecule: yjeq protein;PDBTitle: crystal structure of yjeq from aquifex aeolicus
94 d1bbua1
not modelled
36.4
20
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain95 d1zyma2
not modelled
35.8
13
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system96 d2pxrc1
not modelled
35.5
17
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain97 d1bcce2
not modelled
34.4
9
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor98 c2ivwA_
not modelled
34.4
20
PDB header: lipoproteinChain: A: PDB Molecule: pilp pilot protein;PDBTitle: the solution structure of a domain from the neisseria2 meningitidis pilp pilot protein.
99 c2fb0A_
not modelled
34.3
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of conserved protein of unknown function from2 bacteroides thetaiotaomicron vpi-5482 at 2.10 a resolution, possible3 oxidoreductase
100 d1zlqa1
not modelled
34.2
20
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like101 c2zkrq_
not modelled
33.8
31
PDB header: ribosomal protein/rnaChain: Q: PDB Molecule: rna expansion segment es31 part ii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
102 c3t66A_
not modelled
33.5
13
PDB header: transport proteinChain: A: PDB Molecule: nickel abc transporter (nickel-binding protein);PDBTitle: crystal structure of nickel abc transporter from bacillus halodurans
103 d1x6oa1
not modelled
33.4
17
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: eIF5a N-terminal domain-like104 d1vgya2
not modelled
33.3
17
Fold: Ferredoxin-likeSuperfamily: Bacterial exopeptidase dimerisation domainFamily: Bacterial exopeptidase dimerisation domain105 d1q40b_
not modelled
33.2
26
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: NTF2-like106 d1g7sa1
not modelled
33.1
5
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors107 d1c99a_
not modelled
32.8
39
Fold: Transmembrane helix hairpinSuperfamily: F1F0 ATP synthase subunit CFamily: F1F0 ATP synthase subunit C108 c3c5oD_
not modelled
32.5
18
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: upf0311 protein rpa1785;PDBTitle: crystal structure of the conserved protein of unknown function rpa17852 from rhodopseudomonas palustris
109 c1l6nA_
not modelled
32.4
18
PDB header: viral proteinChain: A: PDB Molecule: gag polyprotein;PDBTitle: structure of the n-terminal 283-residue fragment of the hiv-2 1 gag polyprotein
110 d2plia1
not modelled
32.4
39
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like111 c2l66B_
not modelled
31.8
32
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, abrb family;PDBTitle: the dna-recognition fold of sso7c4 suggests a new member of spovt-abrb2 superfamily from archaea.
112 d1yeza1
not modelled
31.8
10
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TRAM domain113 c2lc4A_
not modelled
31.8
28
PDB header: structural proteinChain: A: PDB Molecule: pilp protein;PDBTitle: solution structure of pilp from pseudomonas aeruginosa
114 c1s1iQ_
not modelled
31.4
31
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l21-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
115 d2hi6a1
not modelled
31.2
41
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: AF0055-like116 d1ixda_
not modelled
31.2
17
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain117 d2if6a1
not modelled
31.0
21
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: YiiX-like118 d3d3ra1
not modelled
30.8
25
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like119 d2a7ya1
not modelled
30.0
19
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Rv2302-like120 c2a7yA_
not modelled
30.0
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein rv2302/mt2359;PDBTitle: solution structure of the conserved hypothetical protein2 rv2302 from the bacterium mycobacterium tuberculosis