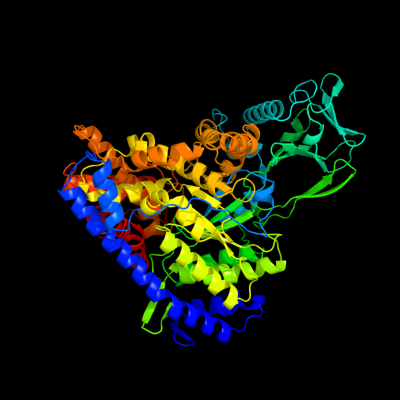
| 1 | d1d8ca_
|
|
 |
100.0 |
97 |
Fold:TIM beta/alpha-barrel
Superfamily:Malate synthase G
Family:Malate synthase G |
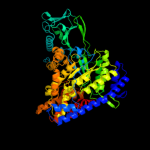
| 2 | d1n8ia_
|
|
 |
100.0 |
60 |
Fold:TIM beta/alpha-barrel
Superfamily:Malate synthase G
Family:Malate synthase G |
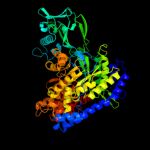
| 3 | c3cuzA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:malate synthase a;
PDBTitle: atomic resolution structures of escherichia coli and2 bacillis anthracis malate synthase a: comparison with3 isoform g and implications for structure based drug design
|
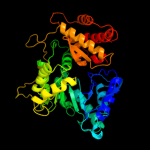
| 4 | c3cuxA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:malate synthase;
PDBTitle: atomic resolution structures of escherichia coli and2 bacillis anthracis malate synthase a: comparison with3 isoform g and implications for structure based drug design
|
| 5 | c3pugA_
|
|
 |
99.8 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:malate synthase;
PDBTitle: haloferax volcanii malate synthase native at 3mm glyoxylate
|
| 6 | c3r4iB_
|
|
 |
99.7 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:citrate lyase;
PDBTitle: crystal structure of a citrate lyase (bxe_b2899) from burkholderia2 xenovorans lb400 at 2.24 a resolution
|
| 7 | c1sgjB_
|
|
 |
99.6 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:citrate lyase, beta subunit;
PDBTitle: crystal structure of citrate lyase beta subunit
|
| 8 | c3qqwC_
|
|
 |
99.6 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:putative citrate lyase;
PDBTitle: crystal structure of a hypothetical lyase (reut_b4148) from ralstonia2 eutropha jmp134 at 2.44 a resolution
|
| 9 | d1sgja_
|
|
 |
99.6 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
| 10 | c1u5vA_
|
|
 |
99.6 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:cite;
PDBTitle: structure of cite complexed with triphosphate group of atp2 form mycobacterium tuberculosis
|
| 11 | d1u5ha_
|
|
 |
99.6 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
| 12 | c2hroA_
|
|
 |
97.0 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of the full-lenght enzyme i of the pts system from2 staphylococcus carnosus
|
| 13 | c2bg5C_
|
|
 |
96.8 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:phosphoenolpyruvate-protein kinase;
PDBTitle: crystal structure of the phosphoenolpyruvate-binding enzyme2 i-domain from the thermoanaerobacter tengcongensis pep:3 sugar phosphotransferase system (pts)
|
| 14 | d1dxea_
|
|
 |
96.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
| 15 | c2hwgA_
|
|
 |
96.5 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate-protein phosphotransferase;
PDBTitle: structure of phosphorylated enzyme i of the2 phosphoenolpyruvate:sugar phosphotransferase system
|
| 16 | c2v5jB_
|
|
 |
96.0 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase;
PDBTitle: apo class ii aldolase hpch
|
| 17 | c3qz6A_
|
|
 |
96.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:hpch/hpai aldolase;
PDBTitle: the crystal structure of hpch/hpai aldolase from desulfitobacterium2 hafniense dcb-2
|
| 18 | c2vwtA_
|
|
 |
95.9 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:yfau, 2-keto-3-deoxy sugar aldolase;
PDBTitle: crystal structure of yfau, a metal ion dependent class ii2 aldolase from escherichia coli k12 - mg-pyruvate product3 complex
|
| 19 | c2olsA_
|
|
 |
93.4 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoenolpyruvate synthase;
PDBTitle: the crystal structure of the phosphoenolpyruvate synthase from2 neisseria meningitidis
|
| 20 | c1izcA_
|
|
 |
93.1 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:macrophomate synthase intermolecular diels-alderase;
PDBTitle: crystal structure analysis of macrophomate synthase
|
| 21 | d1izca_ |
|
not modelled |
93.1 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:HpcH/HpaI aldolase |
| 22 | d1e0ta2 |
|
not modelled |
93.0 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 23 | d1vbga1 |
|
not modelled |
92.8 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 24 | c1h6zA_ |
|
not modelled |
88.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate phosphate dikinase;
PDBTitle: 3.0 a resolution crystal structure of glycosomal pyruvate2 phosphate dikinase from trypanosoma brucei
|
| 25 | d1kbla1 |
|
not modelled |
88.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 26 | d1pkla2 |
|
not modelled |
88.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 27 | d1h6za1 |
|
not modelled |
86.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate phosphate dikinase, C-terminal domain |
| 28 | c1vbhA_ |
|
not modelled |
85.7 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate,orthophosphate dikinase;
PDBTitle: pyruvate phosphate dikinase with bound mg-pep from maize
|
| 29 | d1a3xa2 |
|
not modelled |
81.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 30 | c1kblA_ |
|
not modelled |
74.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate phosphate dikinase;
PDBTitle: pyruvate phosphate dikinase
|
| 31 | d1liua2 |
|
not modelled |
62.5 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 32 | d2g50a2 |
|
not modelled |
59.8 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
| 33 | c2kz6A_ |
|
not modelled |
59.1 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of protein cv0426 from chromobacterium violaceum,2 northeast structural genomics consortium (nesg) target cvt2
|
| 34 | d1njra_ |
|
not modelled |
48.1 |
17 |
Fold:Macro domain-like
Superfamily:Macro domain-like
Family:Macro domain |
| 35 | c2zzxD_ |
|
not modelled |
45.9 |
12 |
PDB header:transport protein
Chain: D: PDB Molecule:abc transporter, solute-binding protein;
PDBTitle: crystal structure of a periplasmic substrate binding protein in2 complex with lactate
|
| 36 | d1zela1 |
|
not modelled |
43.8 |
32 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Rv2827c N-terminal domain-like |
| 37 | c2q4oA_ |
|
not modelled |
39.1 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein at2g37210/t2n18.3;
PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at2g37210
|
| 38 | d2q4oa1 |
|
not modelled |
39.1 |
17 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
| 39 | d1rg6a_ |
|
not modelled |
37.5 |
30 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:SAM (sterile alpha motif) domain |
| 40 | c1zelA_ |
|
not modelled |
36.7 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein rv2827c;
PDBTitle: crystal structure of rv2827c protein from mycobacterium tuberculosis
|
| 41 | d1ic8a2 |
|
not modelled |
35.1 |
27 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:POU-specific domain |
| 42 | c2gbbA_ |
|
not modelled |
31.2 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:putative chorismate mutase;
PDBTitle: crystal structure of secreted chorismate mutase from2 yersinia pestis
|
| 43 | c1zr5B_ |
|
not modelled |
29.7 |
24 |
PDB header:gene regulation
Chain: B: PDB Molecule:h2afy protein;
PDBTitle: crystal structure of the macro-domain of human core histone variant2 macroh2a1.2
|
| 44 | c3q3jA_ |
|
not modelled |
28.2 |
23 |
PDB header:membrane protein/protein binding
Chain: A: PDB Molecule:plexin-a2;
PDBTitle: crystal structure of plexin a2 rbd in complex with rnd1
|
| 45 | d1s9va2 |
|
not modelled |
26.4 |
22 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 46 | d1t35a_ |
|
not modelled |
25.1 |
10 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
| 47 | c3q71A_ |
|
not modelled |
23.6 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:poly [adp-ribose] polymerase 14;
PDBTitle: human parp14 (artd8) - macro domain 2 in complex with adenosine-5-2 diphosphoribose
|
| 48 | d1ydha_ |
|
not modelled |
23.5 |
12 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
| 49 | d1rh6a_ |
|
not modelled |
23.3 |
21 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:Excisionase-like |
| 50 | d1w0ba_ |
|
not modelled |
21.7 |
21 |
Fold:Spectrin repeat-like
Superfamily:Alpha-hemoglobin stabilizing protein AHSP
Family:Alpha-hemoglobin stabilizing protein AHSP |
| 51 | d1muja2 |
|
not modelled |
21.7 |
34 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 52 | c3rmrA_ |
|
not modelled |
19.9 |
33 |
PDB header:protein binding
Chain: A: PDB Molecule:avirulence protein;
PDBTitle: crystal structure of hyaloperonospora arabidopsidis atr1 effector2 domain
|
| 53 | d1jk8a2 |
|
not modelled |
19.9 |
24 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 54 | d1vj7a2 |
|
not modelled |
18.9 |
26 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:RelA/SpoT domain |
| 55 | c2q4dB_ |
|
not modelled |
18.7 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:lysine decarboxylase-like protein at5g11950;
PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at5g11950
|
| 56 | c2zc2A_ |
|
not modelled |
18.3 |
12 |
PDB header:replication
Chain: A: PDB Molecule:dnad-like replication protein;
PDBTitle: crystal structure of dnad-like replication protein from2 streptococcus mutans ua159, gi 24377835, residues 127-199
|
| 57 | d1es0a2 |
|
not modelled |
17.9 |
26 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 58 | d2jn4a1 |
|
not modelled |
17.8 |
23 |
Fold:NifT/FixU barrel-like
Superfamily:NifT/FixU-like
Family:NifT/FixU |
| 59 | c2jn4A_ |
|
not modelled |
17.8 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein fixu, nift;
PDBTitle: solution nmr structure of protein rp4601 from2 rhodopseudomonas palustris. northeast structural genomics3 consortium target rpt2; ontario center for structural4 proteomics target rp4601.
|
| 60 | c2pxgA_ |
|
not modelled |
17.3 |
17 |
PDB header:membrane protein
Chain: A: PDB Molecule:outer membrane protein;
PDBTitle: nmr solution structure of omla
|
| 61 | d1dxsa_ |
|
not modelled |
17.1 |
32 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:SAM (sterile alpha motif) domain |
| 62 | d3bzra1 |
|
not modelled |
16.6 |
25 |
Fold:EscU C-terminal domain-like
Superfamily:EscU C-terminal domain-like
Family:EscU C-terminal domain-like |
| 63 | c3bzrA_ |
|
not modelled |
16.6 |
25 |
PDB header:membrane protein, protein transport
Chain: A: PDB Molecule:escu;
PDBTitle: crystal structure of escu c-terminal domain with n262d mutation, space2 group p 41 21 2
|
| 64 | c3bd1B_ |
|
not modelled |
16.3 |
28 |
PDB header:transcription
Chain: B: PDB Molecule:cro protein;
PDBTitle: structure of the cro protein from putative prophage element xfaso 1 in2 xylella fastidiosa strain ann-1
|
| 65 | c3hlzA_ |
|
not modelled |
16.2 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein bt_1490;
PDBTitle: crystal structure of bt_1490 (np_810393.1) from bacteroides2 thetaiotaomicron vpi-5482 at 1.50 a resolution
|
| 66 | d1mvla_ |
|
not modelled |
15.9 |
26 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
| 67 | c1mvlA_ |
|
not modelled |
15.9 |
26 |
PDB header:lyase
Chain: A: PDB Molecule:ppc decarboxylase athal3a;
PDBTitle: ppc decarboxylase mutant c175s
|
| 68 | c3g80B_ |
|
not modelled |
15.7 |
29 |
PDB header:viral protein
Chain: B: PDB Molecule:protein b2;
PDBTitle: nodamura virus protein b2, rna-binding domain
|
| 69 | c2vy2A_ |
|
not modelled |
15.7 |
50 |
PDB header:transcription
Chain: A: PDB Molecule:protein leafy;
PDBTitle: structure of leafy transcription factor from arabidopsis2 thaliana in complex with dna from ag-i promoter
|
| 70 | c2dnrA_ |
|
not modelled |
15.5 |
15 |
PDB header:rna binding protein
Chain: A: PDB Molecule:synaptojanin-1;
PDBTitle: solution structure of rna binding domain in synaptojanin 1
|
| 71 | c3odmE_ |
|
not modelled |
15.0 |
17 |
PDB header:lyase
Chain: E: PDB Molecule:phosphoenolpyruvate carboxylase;
PDBTitle: archaeal-type phosphoenolpyruvate carboxylase
|
| 72 | c3ogfA_ |
|
not modelled |
14.7 |
14 |
PDB header:de novo protein
Chain: A: PDB Molecule:de novo designed dimeric trefoil-fold sub-domain which
PDBTitle: crystal structure of difoil-4p homo-trimer: de novo designed dimeric2 trefoil-fold sub-domain which forms homo-trimer assembly
|
| 73 | d2ipqx1 |
|
not modelled |
14.2 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:STY4665 C-terminal domain-like |
| 74 | d2ffca1 |
|
not modelled |
14.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
| 75 | d1gsma1 |
|
not modelled |
13.7 |
34 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Immunoglobulin
Family:I set domains |
| 76 | d1ev7a_ |
|
not modelled |
13.1 |
29 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Restriction endonuclease NaeI |
| 77 | d1wuua1 |
|
not modelled |
13.0 |
18 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:GHMP Kinase, N-terminal domain |
| 78 | d1zbsa1 |
|
not modelled |
12.7 |
16 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
| 79 | d1u04a1 |
|
not modelled |
12.6 |
80 |
Fold:SH3-like barrel
Superfamily:PAZ domain
Family:PAZ domain |
| 80 | d1k82a1 |
|
not modelled |
12.2 |
10 |
Fold:S13-like H2TH domain
Superfamily:S13-like H2TH domain
Family:Middle domain of MutM-like DNA repair proteins |
| 81 | d2v9va2 |
|
not modelled |
11.9 |
33 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal fragment of elongation factor SelB |
| 82 | d1ei7a_ |
|
not modelled |
11.8 |
12 |
Fold:Four-helical up-and-down bundle
Superfamily:TMV-like viral coat proteins
Family:TMV-like viral coat proteins |
| 83 | c3da5A_ |
|
not modelled |
11.1 |
67 |
PDB header:rna binding protein
Chain: A: PDB Molecule:argonaute;
PDBTitle: crystal structure of piwi/argonaute/zwille(paz) domain from2 thermococcus thioreducens
|
| 84 | c1pprO_ |
|
not modelled |
10.9 |
23 |
PDB header:light-harvesting protein
Chain: O: PDB Molecule:peridinin-chlorophyll protein;
PDBTitle: peridinin-chlorophyll-protein of amphidinium carterae
|
| 85 | c3ofgA_ |
|
not modelled |
10.8 |
25 |
PDB header:chaperone
Chain: A: PDB Molecule:boca/mesd chaperone for ywtd beta-propeller-egf protein 1;
PDBTitle: structured domain of caenorhabditis elegans bmy-1
|
| 86 | d1qasa3 |
|
not modelled |
10.7 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:PLC-like phosphodiesterases
Family:Mammalian PLC |
| 87 | c2vx6A_ |
|
not modelled |
10.4 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:cellvibrio japonicus mannanase cjman26c;
PDBTitle: cellvibrio japonicus mannanase cjman26c gal1man4-bound form
|
| 88 | d1sv0c_ |
|
not modelled |
10.0 |
18 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:Pointed domain |
| 89 | d1umya_ |
|
not modelled |
10.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
| 90 | d1ykhb1 |
|
not modelled |
9.7 |
19 |
Fold:Mediator hinge subcomplex-like
Superfamily:Mediator hinge subcomplex-like
Family:CSE2-like |
| 91 | d1pava_ |
|
not modelled |
9.7 |
16 |
Fold:IF3-like
Superfamily:SirA-like
Family:SirA-like |
| 92 | d1xzpa3 |
|
not modelled |
9.7 |
35 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:TrmE formyl-THF-binding domain |
| 93 | c1xzqB_ |
|
not modelled |
9.7 |
35 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable trna modification gtpase trme;
PDBTitle: structure of the gtp-binding protein trme from thermotoga2 maritima complexed with 5-formyl-thf
|
| 94 | c3k6qB_ |
|
not modelled |
9.6 |
27 |
PDB header:ligand binding protein
Chain: B: PDB Molecule:putative ligand binding protein;
PDBTitle: crystal structure of an antitoxin part of a putative toxin/antitoxin2 system (swol_0700) from syntrophomonas wolfei subsp. wolfei at 1.80 a3 resolution
|
| 95 | c3cwyA_ |
|
not modelled |
9.5 |
38 |
PDB header:unknown function
Chain: A: PDB Molecule:protein cagd;
PDBTitle: structure of cagd from h. pylori pathogenicity island2 crystallized in the presence of cu(ii) ions
|
| 96 | d1alna1 |
|
not modelled |
9.5 |
16 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Cytidine deaminase |
| 97 | c3c01H_ |
|
not modelled |
9.4 |
10 |
PDB header:membrane protein, protein transport
Chain: H: PDB Molecule:surface presentation of antigens protein spas;
PDBTitle: crystal structural of native spas c-terminal domain
|
| 98 | d1ug3a1 |
|
not modelled |
9.4 |
15 |
Fold:alpha-alpha superhelix
Superfamily:ARM repeat
Family:MIF4G domain-like |
| 99 | c3czoD_ |
|
not modelled |
9.3 |
17 |
PDB header:lyase
Chain: D: PDB Molecule:histidine ammonia-lyase;
PDBTitle: crystal structure of double mutant phenylalanine ammonia-2 lyase from anabaena variabilis
|