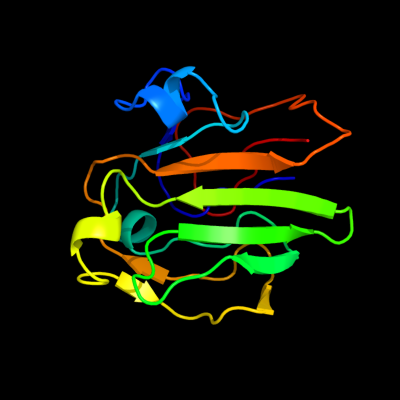
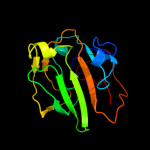
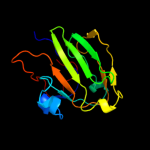
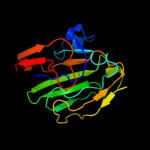
1 d2gpra_
97.4
17
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like2 d1gpra_
96.7
17
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like3 d2f3ga_
96.6
16
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like4 d1glaf_
96.4
13
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like5 c2dncA_
89.6
16
PDB header: transferaseChain: A: PDB Molecule: pyruvate dehydrogenase protein x component;PDBTitle: solution structure of rsgi ruh-054, a lipoyl domain from2 human 2-oxoacid dehydrogenase
6 c2ejmA_
88.8
9
PDB header: ligaseChain: A: PDB Molecule: methylcrotonoyl-coa carboxylase subunit alpha;PDBTitle: solution structure of ruh-072, an apo-biotnyl domain form2 human acetyl coenzyme a carboxylase
7 c2dn8A_
88.7
9
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: solution structure of rsgi ruh-053, an apo-biotin carboxy2 carrier protein from human transcarboxylase
8 d1bdoa_
88.4
12
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains9 c2b8gA_
88.2
13
PDB header: biosynthetic proteinChain: A: PDB Molecule: biotin/lipoyl attachment protein;PDBTitle: solution structure of bacillus subtilis blap biotinylated-2 form (energy minimized mean structure)
10 d1laba_
87.1
11
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains11 c2ejgD_
87.0
10
PDB header: ligaseChain: D: PDB Molecule: 149aa long hypothetical methylmalonyl-coa decarboxylasePDBTitle: crystal structure of the biotin protein ligase (mutation r48a) and2 biotin carboxyl carrier protein complex from pyrococcus horikoshii3 ot3
12 d1y8ob1
85.9
13
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains13 d1dcza_
85.9
11
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains14 d1ghja_
85.1
12
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains15 c2dneA_
84.6
12
PDB header: transferaseChain: A: PDB Molecule: dihydrolipoyllysine-residue acetyltransferasePDBTitle: solution structure of rsgi ruh-058, a lipoyl domain of2 human 2-oxoacid dehydrogenase
16 c2qikA_
83.5
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0131 protein ykqa;PDBTitle: crystal structure of ykqa from bacillus subtilis. northeast2 structural genomics target sr631
17 c2l5tA_
81.8
13
PDB header: transferaseChain: A: PDB Molecule: lipoamide acyltransferase;PDBTitle: solution nmr structure of e2 lipoyl domain from thermoplasma2 acidophilum
18 d1qjoa_
81.7
7
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains19 d1iyua_
81.5
7
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains20 d1gjxa_
81.3
9
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains21 c2kccA_
not modelled
78.9
8
PDB header: ligaseChain: A: PDB Molecule: acetyl-coa carboxylase 2;PDBTitle: solution structure of biotinoyl domain from human acetyl-2 coa carboxylase 2
22 d1k8ma_
not modelled
77.8
10
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains23 c2q8iB_
not modelled
75.7
14
PDB header: transferaseChain: B: PDB Molecule: dihydrolipoyllysine-residue acetyltransferase component ofPDBTitle: pyruvate dehydrogenase kinase isoform 3 in complex with antitumor drug2 radicicol
24 d1pmra_
not modelled
75.4
7
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains25 d1o78a_
not modelled
73.1
9
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains26 c1urzC_
not modelled
59.5
34
PDB header: virus/viral proteinChain: C: PDB Molecule: envelope protein;PDBTitle: low ph induced, membrane fusion conformation of the2 envelope protein of tick-borne encephalitis virus
27 c3cdxB_
not modelled
50.4
13
PDB header: hydrolaseChain: B: PDB Molecule: succinylglutamatedesuccinylase/aspartoacylase;PDBTitle: crystal structure of2 succinylglutamatedesuccinylase/aspartoacylase from3 rhodobacter sphaeroides
28 d1z66a1
not modelled
45.9
22
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain29 c2jqmA_
not modelled
40.9
31
PDB header: transferaseChain: A: PDB Molecule: envelope protein e;PDBTitle: yellow fever envelope protein domain iii nmr structure2 (s288-k398)
30 c3na6A_
not modelled
39.7
19
PDB header: hydrolaseChain: A: PDB Molecule: succinylglutamate desuccinylase/aspartoacylase;PDBTitle: crystal structure of a succinylglutamate desuccinylase (tm1040_2694)2 from silicibacter sp. tm1040 at 2.00 a resolution
31 c1vdzA_
not modelled
39.5
17
PDB header: hydrolaseChain: A: PDB Molecule: a-type atpase subunit a;PDBTitle: crystal structure of a-type atpase catalytic subunit a from2 pyrococcus horikoshii ot3
32 c2hsiB_
not modelled
38.5
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative peptidase m23;PDBTitle: crystal structure of putative peptidase m23 from2 pseudomonas aeruginosa, new york structural genomics3 consortium
33 c2h0pA_
not modelled
38.4
28
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: nmr structure of the dengue-4 virus envelope protein domain2 iii
34 c2qj8B_
not modelled
37.5
11
PDB header: hydrolaseChain: B: PDB Molecule: mlr6093 protein;PDBTitle: crystal structure of an aspartoacylase family protein (mlr6093) from2 mesorhizobium loti maff303099 at 2.00 a resolution
35 d1svba1
not modelled
36.9
22
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain36 d1pjwa_
not modelled
30.2
31
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain37 d1xpma2
not modelled
30.2
40
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like38 c2ka7A_
not modelled
29.7
30
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: nmr solution structure of tm0212 at 40 c
39 d1qwya_
not modelled
29.6
20
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Peptidoglycan hydrolase LytM40 c3mxuA_
not modelled
29.5
20
PDB header: oxidoreductaseChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: crystal structure of glycine cleavage system protein h from bartonella2 henselae
41 c3lazB_
not modelled
28.5
5
PDB header: lyaseChain: B: PDB Molecule: d-galactarate dehydratase;PDBTitle: the crystal structure of the n-terminal domain of d-2 galactarate dehydratase from escherichia coli cft073
42 c2aujD_
not modelled
28.4
15
PDB header: transferaseChain: D: PDB Molecule: dna-directed rna polymerase beta' chain;PDBTitle: structure of thermus aquaticus rna polymerase beta'-subunit2 insert
43 c1otcB_
not modelled
26.9
35
PDB header: protein/dnaChain: B: PDB Molecule: protein (telomere-binding protein beta subunit);PDBTitle: the o. nova telomere end binding protein complexed with2 single strand dna
44 c2i0qB_
not modelled
26.5
35
PDB header: structural protein/dnaChain: B: PDB Molecule: telomere-binding protein beta subunit;PDBTitle: crystal structure of a telomere single-strand dna-protein2 complex from o. nova with full-length alpha and beta3 telomere proteins
45 d1jb7b_
not modelled
26.5
35
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB46 d1ok8a1
not modelled
26.0
25
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain47 c3egpA_
not modelled
25.3
25
PDB header: viral proteinChain: A: PDB Molecule: envelope protein;PDBTitle: crystal structure analysis of dengue-1 envelope protein2 domain iii
48 c2jkuA_
not modelled
24.7
13
PDB header: ligaseChain: A: PDB Molecule: propionyl-coa carboxylase alpha chain,PDBTitle: crystal structure of the n-terminal region of the biotin2 acceptor domain of human propionyl-coa carboxylase
49 c2aukA_
not modelled
24.0
23
PDB header: transferaseChain: A: PDB Molecule: dna-directed rna polymerase beta' chain;PDBTitle: structure of e. coli rna polymerase beta' g/g' insert
50 c2b44A_
not modelled
23.8
20
PDB header: hydrolaseChain: A: PDB Molecule: glycyl-glycine endopeptidase lytm;PDBTitle: truncated s. aureus lytm, p 32 2 1 crystal form
51 c3it5B_
not modelled
22.4
18
PDB header: hydrolaseChain: B: PDB Molecule: protease lasa;PDBTitle: crystal structure of the lasa virulence factor from pseudomonas2 aeruginosa
52 c3iftA_
not modelled
21.2
18
PDB header: oxidoreductaseChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: crystal structure of glycine cleavage system protein h from2 mycobacterium tuberculosis, using x-rays from the compact light3 source.
53 c2edgA_
not modelled
20.7
18
PDB header: biosynthetic proteinChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: solution structure of the gcv_h domain from mouse glycine
54 c2r9qD_
not modelled
19.9
22
PDB header: hydrolaseChain: D: PDB Molecule: 2'-deoxycytidine 5'-triphosphate deaminase;PDBTitle: crystal structure of 2'-deoxycytidine 5'-triphosphate deaminase from2 agrobacterium tumefaciens
55 d1wqla2
not modelled
19.5
24
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Ring hydroxylating alpha subunit catalytic domain56 d1r0va2
not modelled
19.3
11
Fold: Restriction endonuclease-likeSuperfamily: tRNA-intron endonuclease catalytic domain-likeFamily: tRNA-intron endonuclease catalytic domain-like57 c2k33A_
not modelled
19.0
24
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: acra;PDBTitle: solution structure of an n-glycosylated protein using in2 vitro glycosylation
58 d2b1xa2
not modelled
18.2
21
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Ring hydroxylating alpha subunit catalytic domain59 d1onla_
not modelled
18.0
34
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains60 c2gu1A_
not modelled
16.4
16
PDB header: hydrolaseChain: A: PDB Molecule: zinc peptidase;PDBTitle: crystal structure of a zinc containing peptidase from2 vibrio cholerae
61 c3fmcC_
not modelled
15.9
18
PDB header: hydrolaseChain: C: PDB Molecule: putative succinylglutamate desuccinylase / aspartoacylase;PDBTitle: crystal structure of a putative succinylglutamate desuccinylase /2 aspartoacylase family protein (sama_0604) from shewanella amazonensis3 sb2b at 1.80 a resolution
62 d2q07a1
not modelled
15.6
21
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain63 d1ulia2
not modelled
14.8
24
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Ring hydroxylating alpha subunit catalytic domain64 d2bmoa2
not modelled
14.7
16
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Ring hydroxylating alpha subunit catalytic domain65 d1o7na2
not modelled
14.5
16
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Ring hydroxylating alpha subunit catalytic domain66 c3c6dB_
not modelled
13.7
31
PDB header: virusChain: B: PDB Molecule: polyprotein;PDBTitle: the pseudo-atomic structure of dengue immature virus
67 d2f5tx2
not modelled
13.6
21
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: TrmB middle domain-like68 c3lehA_
not modelled
13.5
35
PDB header: transferaseChain: A: PDB Molecule: putative hydroxymethylglutaryl-coa synthase;PDBTitle: the crystal structure of smu.943c from streptococcus mutans ua159
69 c2f9aA_
not modelled
12.9
33
PDB header: transferaseChain: A: PDB Molecule: 3-hydroxy-3-methylglutaryl coenzyme a synthase 1;PDBTitle: hmg-coa synthase from brassica juncea in complex with f-244
70 d1miua3
not modelled
12.5
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB71 c2lc0A_
not modelled
12.4
15
PDB header: protein bindingChain: A: PDB Molecule: putative uncharacterized protein tb39.8;PDBTitle: rv0020c_nter structure
72 d1s6na_
not modelled
12.3
32
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain73 c3k3sG_
not modelled
12.3
15
PDB header: hydrolaseChain: G: PDB Molecule: altronate hydrolase;PDBTitle: crystal structure of altronate hydrolase (fragment 1-84) from shigella2 flexneri.
74 c3csqC_
not modelled
12.3
40
PDB header: hydrolaseChain: C: PDB Molecule: morphogenesis protein 1;PDBTitle: crystal and cryoem structural studies of a cell wall2 degrading enzyme in the bacteriophage phi29 tail
75 c3uajA_
not modelled
12.2
21
PDB header: viral protein/immune systemChain: A: PDB Molecule: envelope protein;PDBTitle: crystal structure of the envelope glycoprotein ectodomain from dengue2 virus serotype 4 in complex with the fab fragment of the chimpanzee3 monoclonal antibody 5h2
76 c3c64A_
not modelled
12.0
25
PDB header: cd36-binding protein,cell adhesionChain: A: PDB Molecule: pfemp1 variant 2 of strain mc;PDBTitle: the mc179 portion of the cysteine-rich interdomain region2 (cidr) of a plasmodium falciparum erythrocyte membrane3 protein-1 (pfemp1)
77 c2f1mA_
not modelled
11.9
15
PDB header: transport proteinChain: A: PDB Molecule: acriflavine resistance protein a;PDBTitle: conformational flexibility in the multidrug efflux system protein acra
78 c1x9eB_
not modelled
11.5
35
PDB header: lyaseChain: B: PDB Molecule: hmg-coa synthase;PDBTitle: crystal structure of hmg-coa synthase from enterococcus2 faecalis
79 d2nqwa1
not modelled
11.5
11
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like80 c1txtB_
not modelled
11.5
40
PDB header: lyaseChain: B: PDB Molecule: 3-hydroxy-3-methylglutaryl-coa synthase;PDBTitle: staphylococcus aureus 3-hydroxy-3-methylglutaryl-coa2 synthase
81 c2wusR_
not modelled
11.3
18
PDB header: structural proteinChain: R: PDB Molecule: putative uncharacterized protein;PDBTitle: bacterial actin mreb assembles in complex with cell shape2 protein rodz
82 c3nyyA_
not modelled
10.9
18
PDB header: hydrolaseChain: A: PDB Molecule: putative glycyl-glycine endopeptidase lytm;PDBTitle: crystal structure of a putative glycyl-glycine endopeptidase lytm2 (rumgna_02482) from ruminococcus gnavus atcc 29149 at 1.60 a3 resolution
83 c2f5tX_
not modelled
10.5
21
PDB header: transcriptionChain: X: PDB Molecule: archaeal transcriptional regulator trmb;PDBTitle: crystal structure of the sugar binding domain of the archaeal2 transcriptional regulator trmb
84 d1ztxe1
not modelled
10.5
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain85 c2p8uB_
not modelled
10.0
43
PDB header: transferaseChain: B: PDB Molecule: hydroxymethylglutaryl-coa synthase, cytoplasmic;PDBTitle: crystal structure of human 3-hydroxy-3-methylglutaryl coa synthase i
86 c3d4rE_
not modelled
9.9
18
PDB header: unknown functionChain: E: PDB Molecule: domain of unknown function from the pfam-b_34464 family;PDBTitle: crystal structure of a duf2118 family protein (mmp0046) from2 methanococcus maripaludis at 2.20 a resolution
87 c1orhA_
not modelled
9.5
14
PDB header: transferaseChain: A: PDB Molecule: protein arginine n-methyltransferase 1;PDBTitle: structure of the predominant protein arginine methyltransferase prmt1
88 c3e9uA_
not modelled
9.1
24
PDB header: membrane proteinChain: A: PDB Molecule: na/ca exchange protein;PDBTitle: crystal structure of calx cbd2 domain
89 d2gtaa1
not modelled
9.0
13
Fold: all-alpha NTP pyrophosphatasesSuperfamily: all-alpha NTP pyrophosphatasesFamily: MazG-like90 c3ddcB_
not modelled
8.9
11
PDB header: hydrolase/apoptosisChain: B: PDB Molecule: ras association domain-containing family protein 5;PDBTitle: crystal structure of nore1a in complex with ras
91 c3n6rK_
not modelled
8.8
16
PDB header: ligaseChain: K: PDB Molecule: propionyl-coa carboxylase, alpha subunit;PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
92 d1iyjb3
not modelled
8.6
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB93 c3fymA_
not modelled
8.3
5
PDB header: dna binding proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: the 1a structure of ymfm, a putative dna-binding membrane2 protein from staphylococcus aureus
94 c3llbA_
not modelled
8.2
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of the protein pa3983 with unknown2 function from pseudomonas aeruginosa pao1
95 c1t5eB_
not modelled
8.1
21
PDB header: transport proteinChain: B: PDB Molecule: multidrug resistance protein mexa;PDBTitle: the structure of mexa
96 d1y7ma2
not modelled
7.7
13
Fold: LysM domainSuperfamily: LysM domainFamily: LysM domain97 d3brda3
not modelled
7.6
13
Fold: beta-TrefoilSuperfamily: DNA-binding protein LAG-1 (CSL)Family: DNA-binding protein LAG-1 (CSL)98 d2a9da1
not modelled
7.4
28
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Molybdenum-containing oxidoreductases-like dimerisation domain99 d2fx0a2
not modelled
7.4
33
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain