| 1 | c3iydD_
|
|
 |
100.0 |
100 |
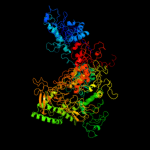
PDB header:transcription/dna
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
|
| 2 | c3h0gA_
|
|
 |
100.0 |
24 |
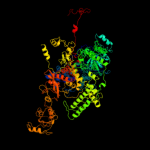
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase ii subunit rpb1;
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
| 3 | d1twfa_
|
|
 |
100.0 |
24 |
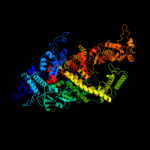
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
| 4 | d1smyd_
|
|
 |
100.0 |
52 |
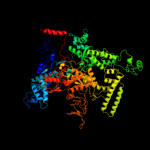
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
| 5 | c1i6hA_
|
|
 |
100.0 |
24 |
PDB header:transcription/dna-rna hybrid
Chain: A: PDB Molecule:dna-directed rna polymerase ii largest subunit;
PDBTitle: rna polymerase ii elongation complex
|
| 6 | d1i6vd_
|
|
 |
100.0 |
51 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
| 7 | c1i6vD_
|
|
 |
100.0 |
51 |
PDB header:transcription
Chain: D: PDB Molecule:dna-directed rna polymerase;
PDBTitle: thermus aquaticus core rna polymerase-rifampicin complex
|
| 8 | d1ynjd1
|
|
 |
100.0 |
58 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
| 9 | c2pmzQ_
|
|
 |
100.0 |
31 |
PDB header:translation, transferase
Chain: Q: PDB Molecule:dna-directed rna polymerase subunit a;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
| 10 | c3qqcA_
|
|
 |
100.0 |
26 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase subunit b, dna-directed rna
PDBTitle: crystal structure of archaeal spt4/5 bound to the rnap clamp domain
|
| 11 | c2pmzG_
|
|
 |
100.0 |
27 |
PDB header:translation, transferase
Chain: G: PDB Molecule:dna-directed rna polymerase subunit a";
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
| 12 | c2aukA_
|
|
 |
98.0 |
98 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: structure of e. coli rna polymerase beta' g/g' insert
|
| 13 | c2aujD_
|
|
 |
97.7 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:dna-directed rna polymerase beta' chain;
PDBTitle: structure of thermus aquaticus rna polymerase beta'-subunit2 insert
|
| 14 | c3e7hA_
|
|
 |
96.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase subunit beta;
PDBTitle: the crystal structure of the beta subunit of the dna-2 directed rna polymerase from vibrio cholerae o1 biovar3 eltor
|
| 15 | c2j7nA_
|
|
 |
89.8 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:rna-dependent rna polymerase;
PDBTitle: structure of the rnai polymerase from neurospora crassa
|
| 16 | c3ky9B_
|
|
 |
62.5 |
12 |
PDB header:apoptosis
Chain: B: PDB Molecule:proto-oncogene vav;
PDBTitle: autoinhibited vav1
|
| 17 | d1nvmb2
|
|
 |
56.4 |
15 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:GAPDH-like |
| 18 | d2f9yb1
|
|
 |
48.4 |
21 |
Fold:ClpP/crotonase
Superfamily:ClpP/crotonase
Family:Biotin dependent carboxylase carboxyltransferase domain |
| 19 | c2f9yB_
|
|
 |
48.4 |
21 |
PDB header:ligase
Chain: B: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl transferase subunit
PDBTitle: the crystal structure of the carboxyltransferase subunit of acc from2 escherichia coli
|
| 20 | c1nvmB_
|
|
 |
47.0 |
15 |
PDB header:lyase/oxidoreductase
Chain: B: PDB Molecule:acetaldehyde dehydrogenase (acylating);
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
| 21 | d1odha_ |
|
not modelled |
46.7 |
21 |
Fold:GCM domain
Superfamily:GCM domain
Family:GCM domain |
| 22 | c2f9iD_ |
|
not modelled |
45.7 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:acetyl-coenzyme a carboxylase carboxyl
PDBTitle: crystal structure of the carboxyltransferase subunit of acc2 from staphylococcus aureus
|
| 23 | c3hcjB_ |
|
not modelled |
42.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide methionine sulfoxide reductase;
PDBTitle: structure of msrb from xanthomonas campestris (oxidized2 form)
|
| 24 | d1l1da_ |
|
not modelled |
41.1 |
18 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:SelR domain |
| 25 | d1xm0a1 |
|
not modelled |
41.1 |
18 |
Fold:Mss4-like
Superfamily:Mss4-like
Family:SelR domain |
| 26 | c2lcqA_ |
|
not modelled |
40.8 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative toxin vapc6;
PDBTitle: solution structure of the endonuclease nob1 from p.horikoshii
|
| 27 | c3cezA_ |
|
not modelled |
40.8 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase;
PDBTitle: crystal structure of methionine-r-sulfoxide reductase from2 burkholderia pseudomallei
|
| 28 | c2l1uA_ |
|
not modelled |
40.6 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase b2, mitochondrial;
PDBTitle: structure-functional analysis of mammalian msrb2 protein
|
| 29 | c1dvbA_ |
|
not modelled |
40.6 |
21 |
PDB header:electron transport
Chain: A: PDB Molecule:rubrerythrin;
PDBTitle: rubrerythrin
|
| 30 | c3e0mB_ |
|
not modelled |
37.7 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peptide methionine sulfoxide reductase msra/msrb
PDBTitle: crystal structure of fusion protein of msra and msrb
|
| 31 | c3a44D_ |
|
not modelled |
37.2 |
33 |
PDB header:metal binding protein
Chain: D: PDB Molecule:hydrogenase nickel incorporation protein hypa;
PDBTitle: crystal structure of hypa in the dimeric form
|
| 32 | d2dk5a1 |
|
not modelled |
36.6 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:RPO3F domain-like |
| 33 | c2k8dA_ |
|
not modelled |
36.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:peptide methionine sulfoxide reductase msrb;
PDBTitle: solution structure of a zinc-binding methionine sulfoxide reductase
|
| 34 | d2g9ha2 |
|
not modelled |
34.6 |
71 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 35 | c3bbnR_ |
|
not modelled |
34.3 |
17 |
PDB header:ribosome
Chain: R: PDB Molecule:ribosomal protein s18;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 36 | d1s9va2 |
|
not modelled |
33.4 |
57 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 37 | c2vrwB_ |
|
not modelled |
31.7 |
19 |
PDB header:signaling protein
Chain: B: PDB Molecule:proto-oncogene vav;
PDBTitle: critical structural role for the ph and c1 domains of the2 vav1 exchange factor
|
| 38 | d2vnud2 |
|
not modelled |
31.7 |
25 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 39 | c2kc1A_ |
|
not modelled |
31.5 |
22 |
PDB header:structural protein
Chain: A: PDB Molecule:mkiaa1027 protein;
PDBTitle: nmr structure of the f0 domain (residues 0-85) of the talin2 ferm domain
|
| 40 | d2eppa1 |
|
not modelled |
30.9 |
35 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 41 | d1fnga2 |
|
not modelled |
30.3 |
71 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 42 | d1nmla1 |
|
not modelled |
29.8 |
28 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Di-heme cytochrome c peroxidase |
| 43 | c3alrA_ |
|
not modelled |
28.1 |
26 |
PDB header:metal binding protein
Chain: A: PDB Molecule:nanos protein;
PDBTitle: crystal structure of nanos
|
| 44 | d1eb7a1 |
|
not modelled |
28.1 |
28 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Di-heme cytochrome c peroxidase |
| 45 | c3bjiA_ |
|
not modelled |
27.4 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:proto-oncogene vav;
PDBTitle: structural basis of promiscuous guanine nucleotide exchange2 by the t-cell essential vav1
|
| 46 | c2kaoA_ |
|
not modelled |
26.9 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine-r-sulfoxide reductase b1;
PDBTitle: structure of reduced mouse methionine sulfoxide reductase b12 (sec95cys mutant)
|
| 47 | d1x64a2 |
|
not modelled |
26.8 |
27 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 48 | c3mkrB_ |
|
not modelled |
26.7 |
31 |
PDB header:transport protein
Chain: B: PDB Molecule:coatomer subunit alpha;
PDBTitle: crystal structure of yeast alpha/epsilon-cop subcomplex of the copi2 vesicular coat
|
| 49 | d1dw0a_ |
|
not modelled |
26.5 |
15 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:monodomain cytochrome c |
| 50 | d1jk8a2 |
|
not modelled |
26.3 |
50 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 51 | d1ekea_ |
|
not modelled |
26.2 |
24 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Ribonuclease H |
| 52 | d1es0a2 |
|
not modelled |
25.0 |
50 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 53 | d1muja2 |
|
not modelled |
24.9 |
50 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 54 | c3mv2A_ |
|
not modelled |
24.9 |
15 |
PDB header:protein transport
Chain: A: PDB Molecule:coatomer subunit alpha;
PDBTitle: crystal structure of a-cop in complex with e-cop
|
| 55 | c3imkA_ |
|
not modelled |
23.3 |
17 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative molybdenum carrier protein;
PDBTitle: crystal structure of putative molybdenum carrier protein (yp_461806.1)2 from syntrophus aciditrophicus sb at 1.45 a resolution
|
| 56 | d1iqca1 |
|
not modelled |
23.0 |
33 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Di-heme cytochrome c peroxidase |
| 57 | c3t6pA_ |
|
not modelled |
22.8 |
16 |
PDB header:apoptosis
Chain: A: PDB Molecule:baculoviral iap repeat-containing protein 2;
PDBTitle: iap antagonist-induced conformational change in ciap1 promotes e32 ligase activation via dimerization
|
| 58 | d1b12a_ |
|
not modelled |
22.4 |
12 |
Fold:LexA/Signal peptidase
Superfamily:LexA/Signal peptidase
Family:Type 1 signal peptidase |
| 59 | d1x62a1 |
|
not modelled |
22.1 |
33 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 60 | d1klua2 |
|
not modelled |
21.5 |
50 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 61 | d2p24a2 |
|
not modelled |
21.4 |
50 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 62 | d1iaka2 |
|
not modelled |
21.4 |
50 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 63 | d2coba1 |
|
not modelled |
21.4 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Psq domain |
| 64 | c3df1R_ |
|
not modelled |
21.3 |
20 |
PDB header:ribosome
Chain: R: PDB Molecule:30s ribosomal protein s18;
PDBTitle: crystal structure of the bacterial ribosome from escherichia2 coli in complex with hygromycin b. this file contains the3 30s subunit of the first 70s ribosome, with hygromycin b4 bound. the entire crystal structure contains two 70s5 ribosomes.
|
| 65 | c3zyqA_ |
|
not modelled |
21.2 |
23 |
PDB header:signaling
Chain: A: PDB Molecule:hepatocyte growth factor-regulated tyrosine kinase
PDBTitle: crystal structure of the tandem vhs and fyve domains of hepatocyte2 growth factor-regulated tyrosine kinase substrate (hgs-hrs) at 1.483 a resolution
|
| 66 | c2yhoE_ |
|
not modelled |
21.0 |
31 |
PDB header:ligase
Chain: E: PDB Molecule:e3 ubiquitin-protein ligase mylip;
PDBTitle: the idol-ube2d complex mediates sterol-dependent degradation of the2 ldl receptor
|
| 67 | d1zvpa2 |
|
not modelled |
20.0 |
42 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:VC0802-like |
| 68 | d1uvqa2 |
|
not modelled |
19.9 |
50 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
| 69 | d2qalr1 |
|
not modelled |
19.5 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Ribosomal protein S18
Family:Ribosomal protein S18 |
| 70 | c3nijA_ |
|
not modelled |
19.3 |
21 |
PDB header:metal binding protein
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase ubr1;
PDBTitle: the structure of ubr box (hiaa)
|
| 71 | c1nltA_ |
|
not modelled |
19.2 |
25 |
PDB header:protein transport
Chain: A: PDB Molecule:mitochondrial protein import protein mas5;
PDBTitle: the crystal structure of hsp40 ydj1
|
| 72 | c3d33B_ |
|
not modelled |
19.0 |
13 |
PDB header:unknown function
Chain: B: PDB Molecule:domain of unknown function with an immunoglobulin-like
PDBTitle: crystal structure of a duf3244 family protein with an immunoglobulin-2 like beta-sandwich fold (bvu_0276) from bacteroides vulgatus atcc3 8482 at 1.70 a resolution
|
| 73 | c1w8xN_ |
|
not modelled |
19.0 |
57 |
PDB header:virus
Chain: N: PDB Molecule:protein p31;
PDBTitle: structural analysis of prd1
|
| 74 | c3fl2A_ |
|
not modelled |
18.9 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase uhrf1;
PDBTitle: crystal structure of the ring domain of the e3 ubiquitin-2 protein ligase uhrf1
|
| 75 | c2ftcP_ |
|
not modelled |
18.6 |
32 |
PDB header:ribosome
Chain: P: PDB Molecule:mitochondrial ribosomal protein l33 isoform a;
PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
|
| 76 | d1ew4a_ |
|
not modelled |
18.6 |
27 |
Fold:N domain of copper amine oxidase-like
Superfamily:Frataxin/Nqo15-like
Family:Frataxin-like |
| 77 | d1hk8a_ |
|
not modelled |
18.4 |
17 |
Fold:PFL-like glycyl radical enzymes
Superfamily:PFL-like glycyl radical enzymes
Family:Class III anaerobic ribonucleotide reductase NRDD subunit |
| 78 | c1hk8A_ |
|
not modelled |
18.4 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:anaerobic ribonucleotide-triphosphate reductase;
PDBTitle: structural basis for allosteric substrate specificity2 regulation in class iii ribonucleotide reductases:3 nrdd in complex with dgtp
|
| 79 | d2uubr1 |
|
not modelled |
16.5 |
21 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Ribosomal protein S18
Family:Ribosomal protein S18 |
| 80 | c2yu3A_ |
|
not modelled |
16.3 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase iii 39 kda
PDBTitle: solution structure of the domain swapped wingedhelix in dna-2 directed rna polymerase iii 39 kda polypeptide
|
| 81 | c3npeA_ |
|
not modelled |
16.1 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:9-cis-epoxycarotenoid dioxygenase 1, chloroplastic;
PDBTitle: structure of vp14 in complex with oxygen
|
| 82 | c2ysaA_ |
|
not modelled |
16.1 |
29 |
PDB header:metal binding protein
Chain: A: PDB Molecule:retinoblastoma-binding protein 6;
PDBTitle: solution structure of the zinc finger cchc domain from the2 human retinoblastoma-binding protein 6 (retinoblastoma-3 binding q protein 1, rbq-1)
|
| 83 | c2kkeA_ |
|
not modelled |
15.4 |
29 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of a dimeric protein of unknown2 function from methanobacterium thermoautotrophicum,3 northeast structural genomics consortium target tr5
|
| 84 | d1i94r_ |
|
not modelled |
15.4 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Ribosomal protein S18
Family:Ribosomal protein S18 |
| 85 | c2riqA_ |
|
not modelled |
15.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:poly [adp-ribose] polymerase 1;
PDBTitle: crystal structure of the third zinc-binding domain of human parp-1
|
| 86 | d1wiia_ |
|
not modelled |
15.0 |
19 |
Fold:Rubredoxin-like
Superfamily:Zinc beta-ribbon
Family:Putative zinc binding domain |
| 87 | d1gu2a_ |
|
not modelled |
14.9 |
11 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:monodomain cytochrome c |
| 88 | d2fug21 |
|
not modelled |
14.9 |
19 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:NQO2-like |
| 89 | c2kdxA_ |
|
not modelled |
14.7 |
14 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:hydrogenase/urease nickel incorporation protein
PDBTitle: solution structure of hypa protein
|
| 90 | c3fybA_ |
|
not modelled |
14.6 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein of unknown function (duf1244);
PDBTitle: crystal structure of a protein of unknown function (duf1244) from2 alcanivorax borkumensis
|
| 91 | d2dula1 |
|
not modelled |
14.3 |
13 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:TRM1-like |
| 92 | c3fzeA_ |
|
not modelled |
14.2 |
21 |
PDB header:protein binding
Chain: A: PDB Molecule:protein ste5;
PDBTitle: structure of the 'minimal scaffold' (ms) domain of ste52 that cocatalyzes fus3 phosphorylation by ste7
|
| 93 | c2kmaA_ |
|
not modelled |
14.2 |
22 |
PDB header:structural protein
Chain: A: PDB Molecule:talin 1;
PDBTitle: nmr structure of the f0f1 double domain (residues 1-202) of2 the talin ferm domain
|
| 94 | c3p04B_ |
|
not modelled |
14.1 |
9 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized bcr;
PDBTitle: crystal structure of the bcr protein from corynebacterium glutamicum.2 northeast structural genomics consortium target cgr8
|
| 95 | d1jb0j_ |
|
not modelled |
13.9 |
50 |
Fold:Single transmembrane helix
Superfamily:Subunit IX of photosystem I reaction centre, PsaJ
Family:Subunit IX of photosystem I reaction centre, PsaJ |
| 96 | c2kn9A_ |
|
not modelled |
13.9 |
18 |
PDB header:electron transport
Chain: A: PDB Molecule:rubredoxin;
PDBTitle: solution structure of zinc-substituted rubredoxin b (rv3250c) from2 mycobacterium tuberculosis. seattle structural genomics center for3 infectious disease target mytud.01635.a
|
| 97 | d1umua_ |
|
not modelled |
13.8 |
14 |
Fold:LexA/Signal peptidase
Superfamily:LexA/Signal peptidase
Family:LexA-related |
| 98 | d1a8ra_ |
|
not modelled |
13.8 |
12 |
Fold:T-fold
Superfamily:Tetrahydrobiopterin biosynthesis enzymes-like
Family:GTP cyclohydrolase I |
| 99 | c1x65A_ |
|
not modelled |
13.7 |
9 |
PDB header:rna binding protein
Chain: A: PDB Molecule:unr protein;
PDBTitle: solution structure of the third cold-shock domain of the human2 kiaa0885 protein (unr protein)
|
















































































































































































































































































































































































































































































































































































































































































































































































































































































