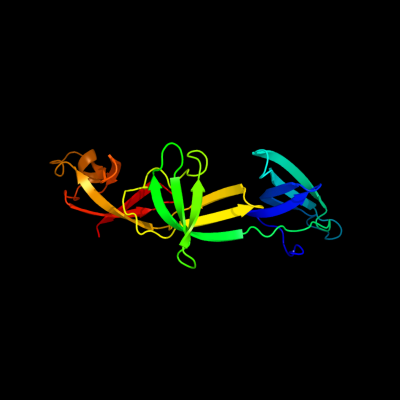
| 1 | c3oyyA_
|
|
 |
100.0 |
30 |
PDB header:translation
Chain: A: PDB Molecule:elongation factor p;
PDBTitle: structure of pseudomonas aeruginosa elongation factor p
|
| 2 | c1uebB_
|
|
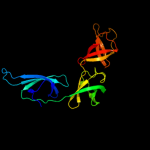 |
100.0 |
31 |
PDB header:rna binding protein
Chain: B: PDB Molecule:elongation factor p;
PDBTitle: crystal structure of translation elongation factor p from2 thermus thermophilus hb8
|
| 3 | c1ybyB_
|
|
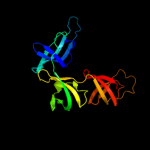 |
100.0 |
29 |
PDB header:translation
Chain: B: PDB Molecule:translation elongation factor p;
PDBTitle: conserved hypothetical protein cth-95 from clostridium2 thermocellum
|
| 4 | c3treA_
|
|
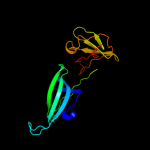 |
100.0 |
23 |
PDB header:translation
Chain: A: PDB Molecule:elongation factor p;
PDBTitle: structure of a translation elongation factor p (efp) from coxiella2 burnetii
|
| 5 | c1iz6B_
|
|
 |
100.0 |
15 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:initiation factor 5a;
PDBTitle: crystal structure of translation initiation factor 5a from pyrococcus2 horikoshii
|
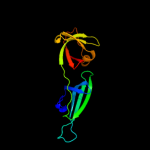
| 6 | c1bkbA_
|
|
 |
100.0 |
19 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor 5a;
PDBTitle: initiation factor 5a from archebacterium pyrobaculum2 aerophilum
|
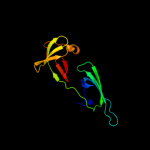
| 7 | c2eifA_
|
|
 |
100.0 |
19 |
PDB header:gene regulation
Chain: A: PDB Molecule:protein (eukaryotic translation initiation factor 5a);
PDBTitle: eukaryotic translation initiation factor 5a from methanococcus2 jannaschii
|
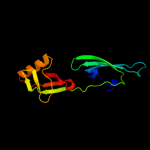
| 8 | c3hksB_
|
|
 |
100.0 |
17 |
PDB header:translation, rna binding protein
Chain: B: PDB Molecule:eukaryotic translation initiation factor 5a-2;
PDBTitle: crystal structure of eukaryotic translation initiation2 factor eif-5a2 from arabidopsis thaliana
|
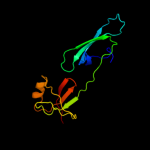
| 9 | c3er0A_
|
|
 |
100.0 |
14 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic translation initiation factor 5a-2;
PDBTitle: crystal structure of the full length eif5a from2 saccharomyces cerevisiae
|
| 10 | c1xtdA_
|
|
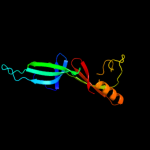 |
99.9 |
17 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic initiation factor 5a;
PDBTitle: structural analysis of leishmania mexicana eukaryotic initiation2 factor 5a
|
| 11 | c3cpfB_
|
|
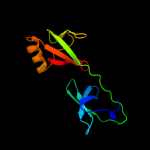 |
99.9 |
13 |
PDB header:cell cycle
Chain: B: PDB Molecule:eukaryotic translation initiation factor 5a-1;
PDBTitle: crystal structure of human eukaryotic translation initiation factor2 eif5a
|
| 12 | d1ueba3
|
|
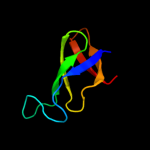 |
99.9 |
47 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 13 | d1ueba1
|
|
 |
99.9 |
22 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 14 | d1ueba2
|
|
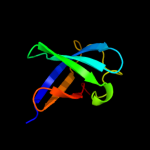 |
99.8 |
25 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 15 | c3a5zF_
|
|
 |
99.8 |
24 |
PDB header:ligase
Chain: F: PDB Molecule:elongation factor p;
PDBTitle: crystal structure of escherichia coli genx in complex with elongation2 factor p
|
| 16 | c1khiA_
|
|
 |
99.3 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:hex1;
PDBTitle: crystal structure of hex1
|
| 17 | d1bkba1
|
|
 |
99.0 |
22 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 18 | d1iz6a1
|
|
 |
99.0 |
16 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 19 | d2eifa1
|
|
 |
99.0 |
17 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 20 | d1x6oa1
|
|
 |
98.4 |
25 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 21 | d1khia1 |
|
not modelled |
97.9 |
18 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 22 | d1bkba2 |
|
not modelled |
97.1 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 23 | d1iz6a2 |
|
not modelled |
96.8 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 24 | d2eifa2 |
|
not modelled |
96.2 |
23 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 25 | d1yiva1 |
|
not modelled |
55.4 |
12 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 26 | c3fp9E_ |
|
not modelled |
47.0 |
22 |
PDB header:hydrolase
Chain: E: PDB Molecule:proteasome-associated atpase;
PDBTitle: crystal structure of intern domain of proteasome-associated2 atpase, mycobacterium tuberculosis
|
| 27 | d1eu1a1 |
|
not modelled |
44.1 |
15 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 28 | d1vlfm1 |
|
not modelled |
43.4 |
20 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 29 | d1g8ka1 |
|
not modelled |
42.5 |
5 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 30 | d1dmra1 |
|
not modelled |
41.7 |
15 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 31 | d2hnxa1 |
|
not modelled |
41.5 |
23 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 32 | d1ogya1 |
|
not modelled |
41.2 |
20 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 33 | d1ftpa_ |
|
not modelled |
39.6 |
20 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 34 | d2iv2x1 |
|
not modelled |
39.2 |
15 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 35 | d2cr5a1 |
|
not modelled |
37.8 |
12 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:UBX domain |
| 36 | d1pmpa_ |
|
not modelled |
37.5 |
11 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 37 | d1kqfa1 |
|
not modelled |
36.3 |
10 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 38 | d2pu9b1 |
|
not modelled |
34.8 |
26 |
Fold:SH3-like barrel
Superfamily:Electron transport accessory proteins
Family:Ferredoxin thioredoxin reductase (FTR), alpha (variable) chain |
| 39 | d1wlfa1 |
|
not modelled |
33.6 |
15 |
Fold:Cdc48 domain 2-like
Superfamily:Cdc48 domain 2-like
Family:Cdc48 domain 2-like |
| 40 | d1y5ia1 |
|
not modelled |
32.7 |
15 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 41 | d2cqaa1 |
|
not modelled |
32.3 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:TIP49 domain |
| 42 | d1slqa_ |
|
not modelled |
30.8 |
14 |
Fold:VP4 membrane interaction domain
Superfamily:VP4 membrane interaction domain
Family:VP4 membrane interaction domain |
| 43 | c1wlfA_ |
|
not modelled |
30.0 |
15 |
PDB header:protein transport
Chain: A: PDB Molecule:peroxisome biogenesis factor 1;
PDBTitle: structure of the n-terminal domain of pex1 aaa-atpase:2 characterization of a putative adaptor-binding domain
|
| 44 | c2z14A_ |
|
not modelled |
29.9 |
16 |
PDB header:signaling protein
Chain: A: PDB Molecule:ef-hand domain-containing family member c2;
PDBTitle: crystal structure of the n-terminal duf1126 in human ef-2 hand domain containing 2 protein
|
| 45 | d2jioa1 |
|
not modelled |
28.7 |
10 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 46 | d1h0ha1 |
|
not modelled |
28.0 |
15 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 47 | c2ki8A_ |
|
not modelled |
27.8 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tungsten formylmethanofuran dehydrogenase,
PDBTitle: solution nmr structure of tungsten formylmethanofuran2 dehydrogenase subunit d from archaeoglobus fulgidus,3 northeast structural genomics consortium target att7
|
| 48 | d1tmoa1 |
|
not modelled |
25.8 |
15 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Formate dehydrogenase/DMSO reductase, C-terminal domain |
| 49 | c1tmoA_ |
|
not modelled |
25.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trimethylamine n-oxide reductase;
PDBTitle: trimethylamine n-oxide reductase from shewanella massilia
|
| 50 | c1h5nC_ |
|
not modelled |
25.4 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dmso reductase;
PDBTitle: dmso reductase modified by the presence of dms and air
|
| 51 | c2dzkA_ |
|
not modelled |
22.8 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ubx domain-containing protein 2;
PDBTitle: structure of the ubx domain in mouse ubx domain-containing2 protein 2
|
| 52 | d1ifca_ |
|
not modelled |
22.7 |
11 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 53 | c3iyuY_ |
|
not modelled |
22.4 |
14 |
PDB header:virus
Chain: Y: PDB Molecule:outer capsid protein vp4;
PDBTitle: atomic model of an infectious rotavirus particle
|
| 54 | c1h0hA_ |
|
not modelled |
22.0 |
15 |
PDB header:dehydrogenase
Chain: A: PDB Molecule:formate dehydrogenase (large subunit);
PDBTitle: tungsten containing formate dehydrogenase from2 desulfovibrio gigas
|
| 55 | c1eu1A_ |
|
not modelled |
20.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dimethyl sulfoxide reductase;
PDBTitle: the crystal structure of rhodobacter sphaeroides dimethylsulfoxide2 reductase reveals two distinct molybdenum coordination environments.
|
| 56 | d1hmsa_ |
|
not modelled |
20.2 |
25 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 57 | d2gp4a1 |
|
not modelled |
20.2 |
19 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:LeuD/IlvD-like
Family:IlvD/EDD C-terminal domain-like |
| 58 | c1g8jC_ |
|
not modelled |
20.0 |
5 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:arsenite oxidase;
PDBTitle: crystal structure analysis of arsenite oxidase from2 alcaligenes faecalis
|
| 59 | c2kxjA_ |
|
not modelled |
18.2 |
14 |
PDB header:protein binding
Chain: A: PDB Molecule:ubx domain-containing protein 4;
PDBTitle: solution structure of ubx domain of human ubxd2 protein
|
| 60 | d1i42a_ |
|
not modelled |
16.4 |
8 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:UBX domain |
| 61 | d2as0a1 |
|
not modelled |
15.7 |
17 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:Hypothetical RNA methyltransferase domain (HRMD) |
| 62 | d1ed7a_ |
|
not modelled |
15.4 |
21 |
Fold:WW domain-like
Superfamily:Carbohydrate binding domain
Family:Carbohydrate binding domain |
| 63 | c3m9bK_ |
|
not modelled |
15.1 |
22 |
PDB header:chaperone
Chain: K: PDB Molecule:proteasome-associated atpase;
PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
|
| 64 | c2ey4A_ |
|
not modelled |
14.9 |
18 |
PDB header:isomerase/biosynthetic protein
Chain: A: PDB Molecule:probable trna pseudouridine synthase b;
PDBTitle: crystal structure of a cbf5-nop10-gar1 complex
|
| 65 | d1mj4a_ |
|
not modelled |
14.9 |
11 |
Fold:Cytochrome b5-like heme/steroid binding domain
Superfamily:Cytochrome b5-like heme/steroid binding domain
Family:Cytochrome b5 |
| 66 | c1ogyA_ |
|
not modelled |
14.5 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:periplasmic nitrate reductase;
PDBTitle: crystal structure of the heterodimeric nitrate reductase2 from rhodobacter sphaeroides
|
| 67 | d2f9ha1 |
|
not modelled |
14.5 |
8 |
Fold:PTSIIA/GutA-like
Superfamily:PTSIIA/GutA-like
Family:PTSIIA/GutA-like |
| 68 | c2iv2X_ |
|
not modelled |
14.5 |
15 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:formate dehydrogenase h;
PDBTitle: reinterpretation of reduced form of formate dehydrogenase h2 from e. coli
|
| 69 | c1vlfQ_ |
|
not modelled |
14.5 |
20 |
PDB header:oxidoreductase
Chain: Q: PDB Molecule:pyrogallol hydroxytransferase large subunit;
PDBTitle: crystal structure of pyrogallol-phloroglucinol2 transhydroxylase from pelobacter acidigallici complexed3 with inhibitor 1,2,4,5-tetrahydroxy-benzene
|
| 70 | d1sa8a_ |
|
not modelled |
14.3 |
11 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 71 | c2wkdA_ |
|
not modelled |
14.3 |
13 |
PDB header:dna binding protein
Chain: A: PDB Molecule:orf34p2;
PDBTitle: crystal structure of a double ile-to-met mutant of protein2 orf34 from lactococcus phage p2
|
| 72 | d1mdca_ |
|
not modelled |
14.2 |
25 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 73 | d1g7na_ |
|
not modelled |
13.9 |
23 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 74 | c1kqgA_ |
|
not modelled |
13.8 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:formate dehydrogenase, nitrate-inducible, major subunit;
PDBTitle: formate dehydrogenase n from e. coli
|
| 75 | c1y5iA_ |
|
not modelled |
13.3 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:respiratory nitrate reductase 1 alpha chain;
PDBTitle: the crystal structure of the narghi mutant nari-k86a
|
| 76 | c2ivfA_ |
|
not modelled |
13.1 |
5 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ethylbenzene dehydrogenase alpha-subunit;
PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
|
| 77 | d1hkoa_ |
|
not modelled |
13.0 |
16 |
Fold:Cytochrome b5-like heme/steroid binding domain
Superfamily:Cytochrome b5-like heme/steroid binding domain
Family:Cytochrome b5 |
| 78 | c2rrfA_ |
|
not modelled |
12.9 |
19 |
PDB header:unknown function
Chain: A: PDB Molecule:zinc finger fyve domain-containing protein 21;
PDBTitle: the solution structure of the c-terminal region of zinc finger fyve2 domain-containing protein 21
|
| 79 | d1opaa_ |
|
not modelled |
12.9 |
18 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 80 | d1fdqa_ |
|
not modelled |
12.9 |
20 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 81 | d2cu8a2 |
|
not modelled |
12.5 |
21 |
Fold:Glucocorticoid receptor-like (DNA-binding domain)
Superfamily:Glucocorticoid receptor-like (DNA-binding domain)
Family:LIM domain |
| 82 | d1kv7a2 |
|
not modelled |
12.5 |
12 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 83 | d1hfua1 |
|
not modelled |
12.4 |
10 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Multidomain cupredoxins |
| 84 | d1kqwa_ |
|
not modelled |
11.8 |
17 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 85 | c2v45A_ |
|
not modelled |
11.7 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:periplasmic nitrate reductase;
PDBTitle: a new catalytic mechanism of periplasmic nitrate reductase2 from desulfovibrio desulfuricans atcc 27774 from3 crystallographic and epr data and based on detailed4 analysis of the sixth ligand
|
| 86 | d1iccc_ |
|
not modelled |
11.6 |
8 |
Fold:Cytochrome b5-like heme/steroid binding domain
Superfamily:Cytochrome b5-like heme/steroid binding domain
Family:Cytochrome b5 |
| 87 | c1cr5B_ |
|
not modelled |
11.4 |
8 |
PDB header:endocytosis/exocytosis
Chain: B: PDB Molecule:sec18p (residues 22 - 210);
PDBTitle: n-terminal domain of sec18p
|
| 88 | c2vq5B_ |
|
not modelled |
11.2 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:s-norcoclaurine synthase;
PDBTitle: x-ray structure of norcoclaurine synthase from thalictrum2 flavum in complex with dopamine and hydroxybenzaldehyde
|
| 89 | d1m2ia_ |
|
not modelled |
11.0 |
16 |
Fold:Cytochrome b5-like heme/steroid binding domain
Superfamily:Cytochrome b5-like heme/steroid binding domain
Family:Cytochrome b5 |
| 90 | c2nyaF_ |
|
not modelled |
10.7 |
15 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:periplasmic nitrate reductase;
PDBTitle: crystal structure of the periplasmic nitrate reductase2 (nap) from escherichia coli
|
| 91 | d1x9la_ |
|
not modelled |
10.6 |
20 |
Fold:Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily:DR1885-like metal-binding protein
Family:DR1885-like metal-binding protein |
| 92 | d1h8ca_ |
|
not modelled |
10.5 |
25 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:UBX domain |
| 93 | c2gp4A_ |
|
not modelled |
10.3 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:6-phosphogluconate dehydratase;
PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
|
| 94 | d1vioa2 |
|
not modelled |
10.3 |
15 |
Fold:Alpha-L RNA-binding motif
Superfamily:Alpha-L RNA-binding motif
Family:Pseudouridine synthase RsuA N-terminal domain |
| 95 | c3e0jG_ |
|
not modelled |
10.1 |
10 |
PDB header:transferase
Chain: G: PDB Molecule:dna polymerase subunit delta-2;
PDBTitle: x-ray structure of the complex of regulatory subunits of2 human dna polymerase delta
|
| 96 | d1bwya_ |
|
not modelled |
10.0 |
20 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 97 | d2ix0a3 |
|
not modelled |
10.0 |
10 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 98 | c2apoA_ |
|
not modelled |
9.8 |
14 |
PDB header:isomerase/rna binding protein
Chain: A: PDB Molecule:probable trna pseudouridine synthase b;
PDBTitle: crystal structure of the methanococcus jannaschii cbf52 nop10 complex
|
| 99 | c2ibjA_ |
|
not modelled |
9.7 |
12 |
PDB header:electron transport
Chain: A: PDB Molecule:cytochrome b5;
PDBTitle: structure of house fly cytochrome b5
|