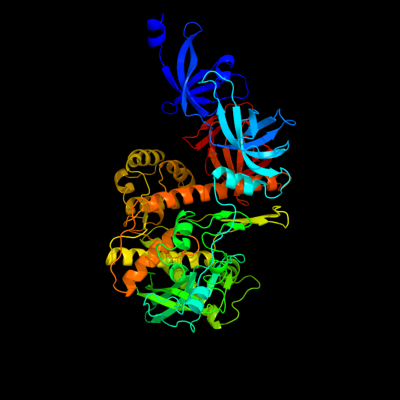
| 1 | c2ix1A_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:exoribonuclease 2;
PDBTitle: rnase ii d209n mutant
|
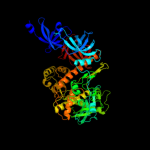
| 2 | c2wp8J_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: J: PDB Molecule:exosome complex exonuclease dis3;
PDBTitle: yeast rrp44 nuclease
|
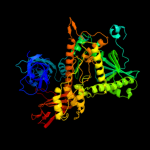
| 3 | c2vnuD_
|
|
 |
100.0 |
25 |
PDB header:hydrolase/rna
Chain: D: PDB Molecule:exosome complex exonuclease rrp44;
PDBTitle: crystal structure of sc rrp44
|
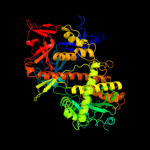
| 4 | c2r7fA_
|
|
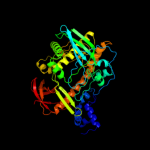 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease ii family protein;
PDBTitle: crystal structure of ribonuclease ii family protein from2 deinococcus radiodurans, hexagonal crystal form. northeast3 structural genomics target drr63
|
| 5 | d2r7da2
|
|
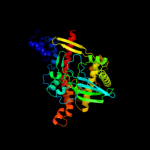 |
100.0 |
26 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:RNB domain-like |
| 6 | d2ix0a4
|
|
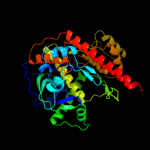 |
100.0 |
28 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:RNB domain-like |
| 7 | d2vnud4
|
|
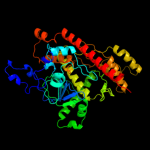 |
100.0 |
27 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:RNB domain-like |
| 8 | d2ix0a1
|
|
 |
99.7 |
15 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 9 | d1go3e1
|
|
 |
99.6 |
30 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 10 | c3h0gS_
|
|
 |
99.6 |
16 |
PDB header:transcription
Chain: S: PDB Molecule:dna-directed rna polymerase ii subunit rpb7;
PDBTitle: rna polymerase ii from schizosaccharomyces pombe
|
| 11 | d2c35b1
|
|
 |
99.6 |
13 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 12 | d2vnud1
|
|
 |
99.5 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 13 | d2ix0a2
|
|
 |
99.4 |
25 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 14 | c2b8kG_
|
|
 |
99.4 |
18 |
PDB header:transferase
Chain: G: PDB Molecule:dna-directed rna polymerase ii 19 kda
PDBTitle: 12-subunit rna polymerase ii
|
| 15 | c2c35F_
|
|
 |
99.4 |
12 |
PDB header:polymerase
Chain: F: PDB Molecule:dna-directed rna polymerase ii 19 kda
PDBTitle: subunits rpb4 and rpb7 of human rna polymerase ii
|
| 16 | c1go3E_
|
|
 |
99.3 |
29 |
PDB header:transferase
Chain: E: PDB Molecule:dna-directed rna polymerase subunit e;
PDBTitle: structure of an archeal homolog of the eukaryotic rna2 polymerase ii rpb4/rpb7 complex
|
| 17 | c2k52A_
|
|
 |
99.2 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein mj1198;
PDBTitle: structure of uncharacterized protein mj1198 from2 methanocaldococcus jannaschii. northeast structural3 genomics target mjr117b
|
| 18 | d1y14b1
|
|
 |
99.2 |
23 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 19 | c2pmzE_
|
|
 |
99.2 |
30 |
PDB header:translation, transferase
Chain: E: PDB Molecule:dna-directed rna polymerase subunit e;
PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
|
| 20 | d2vnud3
|
|
 |
99.2 |
15 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 21 | c1nt9G_ |
|
not modelled |
99.2 |
20 |
PDB header:transcription, transferase
Chain: G: PDB Molecule:dna-directed rna polymerase ii 19 kd polypeptide;
PDBTitle: complete 12-subunit rna polymerase ii
|
| 22 | d1kl9a2 |
|
not modelled |
99.2 |
32 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 23 | c2ckzB_ |
|
not modelled |
99.1 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:dna-directed rna polymerase iii 25 kd
PDBTitle: x-ray structure of rna polymerase iii subcomplex c17-c25.
|
| 24 | c2cqoA_
|
|
 |
99.1 |
20 |
PDB header:ribosome
Chain: A: PDB Molecule:nucleolar protein of 40 kda;
PDBTitle: solution structure of the s1 rna binding domain of human2 hypothetical protein flj11067
|
| 25 | d1q46a2 |
|
not modelled |
99.1 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 26 | c1yz6A_ |
|
not modelled |
99.1 |
22 |
PDB header:translation
Chain: A: PDB Molecule:probable translation initiation factor 2 alpha
PDBTitle: crystal structure of intact alpha subunit of aif2 from2 pyrococcus abyssi
|
| 27 | c1q46A_ |
|
not modelled |
99.1 |
21 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor 2 alpha subunit;
PDBTitle: crystal structure of the eif2 alpha subunit from2 saccharomyces cerevisia
|
| 28 | c2khiA_ |
|
not modelled |
99.0 |
29 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s1;
PDBTitle: nmr structure of the domain 4 of the e. coli ribosomal2 protein s1
|
| 29 | c2khjA_ |
|
not modelled |
99.0 |
26 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s1;
PDBTitle: nmr structure of the domain 6 of the e. coli ribosomal2 protein s1
|
| 30 | c2k4kA_ |
|
not modelled |
98.9 |
35 |
PDB header:rna binding protein
Chain: A: PDB Molecule:general stress protein 13;
PDBTitle: solution structure of gsp13 from bacillus subtilis
|
| 31 | c2eqsA_ |
|
not modelled |
98.9 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:atp-dependent rna helicase dhx8;
PDBTitle: solution structure of the s1 rna binding domain of human2 atp-dependent rna helicase dhx8
|
| 32 | c1q8kA_ |
|
not modelled |
98.9 |
31 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic translation initiation factor 2
PDBTitle: solution structure of alpha subunit of human eif2
|
| 33 | c2oceA_ |
|
not modelled |
98.9 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein pa5201;
PDBTitle: crystal structure of tex family protein pa5201 from2 pseudomonas aeruginosa
|
| 34 | d3bzka4 |
|
not modelled |
98.9 |
33 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 35 | c3psiA_ |
|
not modelled |
98.8 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation factor spt6;
PDBTitle: crystal structure of the spt6 core domain from saccharomyces2 cerevisiae, form spt6(239-1451)
|
| 36 | c2ahoB_ |
|
not modelled |
98.8 |
22 |
PDB header:translation
Chain: B: PDB Molecule:translation initiation factor 2 alpha subunit;
PDBTitle: structure of the archaeal initiation factor eif2 alpha-2 gamma heterodimer from sulfolobus solfataricus complexed3 with gdpnp
|
| 37 | c1kl9A_ |
|
not modelled |
98.8 |
30 |
PDB header:translation
Chain: A: PDB Molecule:eukaryotic translation initiation factor 2 subunit 1;
PDBTitle: crystal structure of the n-terminal segment of human eukaryotic2 initiation factor 2alpha
|
| 38 | d1sroa_ |
|
not modelled |
98.7 |
31 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 39 | d2z0sa1 |
|
not modelled |
98.7 |
19 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 40 | d2ba0a1 |
|
not modelled |
98.7 |
19 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 41 | d2je6i1 |
|
not modelled |
98.7 |
16 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 42 | d2ahob2 |
|
not modelled |
98.6 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 43 | d1wi5a_ |
|
not modelled |
98.6 |
19 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 44 | c1l2fA_ |
|
not modelled |
98.6 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:n utilization substance protein a;
PDBTitle: crystal structure of nusa from thermotoga maritima: a2 structure-based role of the n-terminal domain
|
| 45 | c3go5A_ |
|
not modelled |
98.6 |
25 |
PDB header:gene regulation
Chain: A: PDB Molecule:multidomain protein with s1 rna-binding domains;
PDBTitle: crystal structure of a multidomain protein with nucleic acid binding2 domains (sp_0946) from streptococcus pneumoniae tigr4 at 1.40 a3 resolution
|
| 46 | c1hh2P_ |
|
not modelled |
98.4 |
14 |
PDB header:transcription regulation
Chain: P: PDB Molecule:n utilization substance protein a;
PDBTitle: crystal structure of nusa from thermotoga maritima
|
| 47 | d1hh2p1 |
|
not modelled |
98.4 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 48 | d2nn6h1 |
|
not modelled |
98.3 |
23 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 49 | c3ayhB_ |
|
not modelled |
98.2 |
15 |
PDB header:transcription
Chain: B: PDB Molecule:dna-directed rna polymerase iii subunit rpc8;
PDBTitle: crystal structure of the c17/25 subcomplex from s. pombe rna2 polymerase iii
|
| 50 | c2z0sA_ |
|
not modelled |
98.2 |
19 |
PDB header:rna binding protein
Chain: A: PDB Molecule:probable exosome complex rna-binding protein 1;
PDBTitle: crystal structure of putative exosome complex rna-binding2 protein
|
| 51 | c3k2zA_
|
|
 |
98.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:lexa repressor;
PDBTitle: crystal structure of a lexa protein from thermotoga maritima
|
| 52 | c2bh8B_ |
|
not modelled |
97.8 |
51 |
PDB header:transcription
Chain: B: PDB Molecule:1b11;
PDBTitle: combinatorial protein 1b11
|
| 53 | d2ix0a3 |
|
not modelled |
97.7 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 54 | d2vnud2 |
|
not modelled |
97.7 |
26 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 55 | c2ba0A_ |
|
not modelled |
97.7 |
19 |
PDB header:rna binding protein
Chain: A: PDB Molecule:archeal exosome rna binding protein rrp4;
PDBTitle: archaeal exosome core
|
| 56 | d1smxa_ |
|
not modelled |
97.7 |
18 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 57 | d2nn6i1 |
|
not modelled |
97.6 |
19 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 58 | c3aqqD_ |
|
not modelled |
97.4 |
18 |
PDB header:dna binding protein
Chain: D: PDB Molecule:calcium-regulated heat stable protein 1;
PDBTitle: crystal structure of human crhsp-24
|
| 59 | c2a8vA_ |
|
not modelled |
97.3 |
24 |
PDB header:protein/rna
Chain: A: PDB Molecule:rna binding domain of rho transcription
PDBTitle: rho transcription termination factor/rna complex
|
| 60 | c2je6I_ |
|
not modelled |
97.2 |
17 |
PDB header:hydrolase
Chain: I: PDB Molecule:exosome complex rna-binding protein 1;
PDBTitle: structure of a 9-subunit archaeal exosome
|
| 61 | c2nn6I_ |
|
not modelled |
97.0 |
19 |
PDB header:hydrolase/transferase
Chain: I: PDB Molecule:3'-5' exoribonuclease csl4 homolog;
PDBTitle: structure of the human rna exosome composed of rrp41, rrp45,2 rrp46, rrp43, mtr3, rrp42, csl4, rrp4, and rrp40
|
| 62 | c2rf4A_ |
|
not modelled |
97.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase i subunit rpa4;
PDBTitle: crystal structure of the rna polymerase i subcomplex a14/43
|
| 63 | c3a0jB_ |
|
not modelled |
96.9 |
26 |
PDB header:transcription
Chain: B: PDB Molecule:cold shock protein;
PDBTitle: crystal structure of cold shock protein 1 from thermus2 thermophilus hb8
|
| 64 | d1a62a2 |
|
not modelled |
96.9 |
28 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 65 | d1mjca_ |
|
not modelled |
96.9 |
21 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 66 | c2ytxA_ |
|
not modelled |
96.8 |
24 |
PDB header:rna binding protein
Chain: A: PDB Molecule:cold shock domain-containing protein e1;
PDBTitle: solution structure of the second cold-shock domain of the human2 kiaa0885 protein (unr protein)
|
| 67 | d1g6pa_ |
|
not modelled |
96.8 |
22 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 68 | d1wfqa_ |
|
not modelled |
96.7 |
22 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 69 | d1mkma1
|
|
 |
96.7 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Transcriptional regulator IclR, N-terminal domain |
| 70 | c2ytyA_ |
|
not modelled |
96.6 |
17 |
PDB header:rna binding protein
Chain: A: PDB Molecule:cold shock domain-containing protein e1;
PDBTitle: solution structure of the fourth cold-shock domain of the human2 kiaa0885 protein (unr protein)
|
| 71 | d1h95a_ |
|
not modelled |
96.6 |
25 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 72 | c2ba1B_ |
|
not modelled |
96.6 |
20 |
PDB header:rna binding protein
Chain: B: PDB Molecule:archaeal exosome rna binding protein csl4;
PDBTitle: archaeal exosome core
|
| 73 | c1mkmA_
|
|
 |
96.5 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:iclr transcriptional regulator;
PDBTitle: crystal structure of the thermotoga maritima iclr
|
| 74 | c3camB_ |
|
not modelled |
96.5 |
23 |
PDB header:gene regulation
Chain: B: PDB Molecule:cold-shock domain family protein;
PDBTitle: crystal structure of the cold shock domain protein from neisseria2 meningitidis
|
| 75 | c2k5nA_ |
|
not modelled |
96.3 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative cold-shock protein;
PDBTitle: solution nmr structure of the n-terminal domain of protein2 eca1580 from erwinia carotovora, northeast structural3 genomics consortium target ewr156a
|
| 76 | c2xroE_ |
|
not modelled |
95.9 |
15 |
PDB header:dna-binding protein/dna
Chain: E: PDB Molecule:hth-type transcriptional regulator ttgv;
PDBTitle: crystal structure of ttgv in complex with its dna operator
|
| 77 | d1jhfa1 |
|
not modelled |
95.9 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:LexA repressor, N-terminal DNA-binding domain |
| 78 | c1j5yA_ |
|
not modelled |
95.9 |
19 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, biotin repressor family;
PDBTitle: crystal structure of transcriptional regulator (tm1602) from2 thermotoga maritima at 2.3 a resolution
|
| 79 | c3mq0A_ |
|
not modelled |
95.8 |
16 |
PDB header:transcription repressor
Chain: A: PDB Molecule:transcriptional repressor of the blcabc operon;
PDBTitle: crystal structure of agobacterium tumefaciens repressor blcr
|
| 80 | c3hruA_ |
|
not modelled |
95.6 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:metalloregulator scar;
PDBTitle: crystal structure of scar with bound zn2+
|
| 81 | d2es2a1 |
|
not modelled |
95.5 |
24 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 82 | c2kcmA_ |
|
not modelled |
95.4 |
21 |
PDB header:nucleic acid binding protein
Chain: A: PDB Molecule:cold shock domain family protein;
PDBTitle: solution nmr structure of the n-terminal ob-domain of so_1732 from2 shewanella oneidensis. northeast structural genomics consortium3 target sor210a.
|
| 83 | c1x65A_ |
|
not modelled |
95.2 |
18 |
PDB header:rna binding protein
Chain: A: PDB Molecule:unr protein;
PDBTitle: solution structure of the third cold-shock domain of the human2 kiaa0885 protein (unr protein)
|
| 84 | d1c9oa_ |
|
not modelled |
95.2 |
25 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 85 | d1biaa1 |
|
not modelled |
95.1 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Biotin repressor-like |
| 86 | c2o0yB_
|
|
 |
95.0 |
16 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: crystal structure of putative transcriptional regulator rha1_ro069532 (iclr-family) from rhodococcus sp.
|
| 87 | d1j5ya1 |
|
not modelled |
95.0 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Biotin repressor-like |
| 88 | c1uwvA_ |
|
not modelled |
94.9 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:23s rrna (uracil-5-)-methyltransferase ruma;
PDBTitle: crystal structure of ruma, the iron-sulfur cluster2 containing e. coli 23s ribosomal rna 5-methyluridine3 methyltransferase
|
| 89 | d1xmka1 |
|
not modelled |
94.8 |
22 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Z-DNA binding domain |
| 90 | c2g7uB_ |
|
not modelled |
94.8 |
20 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:transcriptional regulator;
PDBTitle: 2.3 a structure of putative catechol degradative operon regulator from2 rhodococcus sp. rha1
|
| 91 | d1uwva1 |
|
not modelled |
94.8 |
15 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:TRAM domain |
| 92 | d1stza1 |
|
not modelled |
94.6 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Heat-inducible transcription repressor HrcA, N-terminal domain |
| 93 | d2p4wa1 |
|
not modelled |
94.5 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:PF1790-like |
| 94 | d2nn6g1 |
|
not modelled |
94.4 |
15 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 95 | c1z6rC_ |
|
not modelled |
94.3 |
10 |
PDB header:transcription
Chain: C: PDB Molecule:mlc protein;
PDBTitle: crystal structure of mlc from escherichia coli
|
| 96 | d2d1ha1 |
|
not modelled |
94.3 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:TrmB-like |
| 97 | c2it0A_ |
|
not modelled |
94.1 |
24 |
PDB header:transcription/dna
Chain: A: PDB Molecule:iron-dependent repressor ider;
PDBTitle: crystal structure of a two-domain ider-dna complex crystal2 form ii
|
| 98 | d2hr3a1 |
|
not modelled |
94.0 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:MarR-like transcriptional regulators |
| 99 | c1f5tA_ |
|
not modelled |
94.0 |
21 |
PDB header:transcription/dna
Chain: A: PDB Molecule:diphtheria toxin repressor;
PDBTitle: diphtheria tox repressor (c102d mutant) complexed with2 nickel and dtxr consensus binding sequence
|
| 100 | c3trzE_ |
|
not modelled |
93.9 |
19 |
PDB header:rna binding protein/rna
Chain: E: PDB Molecule:protein lin-28 homolog a;
PDBTitle: mouse lin28a in complex with let-7d microrna pre-element
|
| 101 | c2ytvA_ |
|
not modelled |
93.9 |
23 |
PDB header:rna binding protein
Chain: A: PDB Molecule:cold shock domain-containing protein e1;
PDBTitle: solution structure of the fifth cold-shock domain of the human2 kiaa0885 protein (unr protein)
|
| 102 | d1z05a1 |
|
not modelled |
93.8 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:ROK associated domain |
| 103 | d1luza_ |
|
not modelled |
93.7 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 104 | c2oqgA_ |
|
not modelled |
93.7 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:possible transcriptional regulator, arsr family protein;
PDBTitle: arsr-like transcriptional regulator from rhodococcus sp. rha1
|
| 105 | d1lvaa3 |
|
not modelled |
93.3 |
23 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal fragment of elongation factor SelB |
| 106 | c3r4kD_ |
|
not modelled |
93.3 |
14 |
PDB header:dna binding protein
Chain: D: PDB Molecule:transcriptional regulator, iclr family;
PDBTitle: crystal structure of a putative iclr transcriptional regulator2 (tm1040_3717) from silicibacter sp. tm1040 at 2.46 a resolution
|
| 107 | c1k0rB_ |
|
not modelled |
93.1 |
26 |
PDB header:transcription
Chain: B: PDB Molecule:nusa;
PDBTitle: crystal structure of mycobacterium tuberculosis nusa
|
| 108 | c3pqkD_ |
|
not modelled |
93.0 |
26 |
PDB header:transcription
Chain: D: PDB Molecule:biofilm growth-associated repressor;
PDBTitle: crystal structure of the transcriptional repressor bigr from xylella2 fastidiosa
|
| 109 | c3f6vA_ |
|
not modelled |
92.9 |
17 |
PDB header:transcription regulator
Chain: A: PDB Molecule:possible transcriptional regulator, arsr family
PDBTitle: crystal structure of possible transcriptional regulator for2 arsenical resistance
|
| 110 | d2htja1 |
|
not modelled |
92.8 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:FaeA-like |
| 111 | c1wsuA_ |
|
not modelled |
92.8 |
21 |
PDB header:translation/rna
Chain: A: PDB Molecule:selenocysteine-specific elongation factor;
PDBTitle: c-terminal domain of elongation factor selb complexed with2 secis rna
|
| 112 | c1fx7C_ |
|
not modelled |
92.7 |
25 |
PDB header:signaling protein
Chain: C: PDB Molecule:iron-dependent repressor ider;
PDBTitle: crystal structure of the iron-dependent regulator (ider)2 from mycobacterium tuberculosis
|
| 113 | c2h09A_ |
|
not modelled |
92.6 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator mntr;
PDBTitle: crystal structure of diphtheria toxin repressor like protein2 from e. coli
|
| 114 | d1r1ua_ |
|
not modelled |
92.6 |
17 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:ArsR-like transcriptional regulators |
| 115 | d1ku9a_ |
|
not modelled |
92.4 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:DNA-binding protein Mj223 |
| 116 | c2ewnA_ |
|
not modelled |
92.4 |
18 |
PDB header:ligase, transcription
Chain: A: PDB Molecule:bira bifunctional protein;
PDBTitle: ecoli biotin repressor with co-repressor analog
|
| 117 | c1r22B_ |
|
not modelled |
92.2 |
17 |
PDB header:transcription repressor
Chain: B: PDB Molecule:transcriptional repressor smtb;
PDBTitle: crystal structure of the cyanobacterial metallothionein2 repressor smtb (c14s/c61s/c121s mutant) in the zn2alpha5-3 form
|
| 118 | c3cuoB_ |
|
not modelled |
92.0 |
24 |
PDB header:transcription regulator
Chain: B: PDB Molecule:uncharacterized hth-type transcriptional regulator ygav;
PDBTitle: crystal structure of the predicted dna-binding transcriptional2 regulator from e. coli
|
| 119 | c3b73A_ |
|
not modelled |
91.9 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phih1 repressor-like protein;
PDBTitle: crystal structure of the phih1 repressor-like protein from2 haloarcula marismortui
|
| 120 | c2fe3B_ |
|
not modelled |
91.9 |
20 |
PDB header:dna binding protein
Chain: B: PDB Molecule:peroxide operon regulator;
PDBTitle: the crystal structure of bacillus subtilis perr-zn reveals a novel2 zn(cys)4 structural redox switch
|