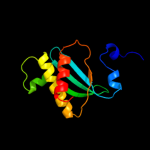
| 1 | d2gycm1 |
|
 |
100.0 |
100 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
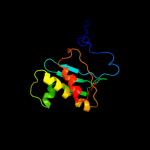
| 2 | c3bboQ_ |
|
 |
100.0 |
46 |
PDB header:ribosome
Chain: Q: PDB Molecule:ribosomal protein l18;
PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
|
| 3 | d1ovya_ |
|
 |
100.0 |
57 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
| 4 | c2jl8S_ |
|
 |
100.0 |
45 |
PDB header:ribosome
Chain: S: PDB Molecule:50s ribosomal protein l18;
PDBTitle: insights into translational termination from the structure2 of rf2 bound to the ribosome (part 4 of 4).3 this file contains the 50s subunit.
|
| 5 | d2zjrl1 |
|
 |
100.0 |
50 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
| 6 | d2j01s1 |
|
 |
100.0 |
47 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
| 7 | d1vqon1 |
|
 |
100.0 |
28 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
| 8 | c2zkrn_ |
|
 |
99.8 |
23 |
PDB header:ribosomal protein/rna
Chain: N: PDB Molecule:rna expansion segment es27;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 9 | c4a1aM_ |
|
 |
99.8 |
27 |
PDB header:ribosome
Chain: M: PDB Molecule:60s ribosomal protein l5;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
|
| 10 | c3izcQ_ |
|
 |
99.8 |
29 |
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein rpl5 (l18p);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 11 | c3iz5Q_ |
|
 |
99.7 |
27 |
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein l5 (l18p);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
|
| 12 | c1s1iE_ |
|
 |
99.6 |
28 |
PDB header:ribosome
Chain: E: PDB Molecule:60s ribosomal protein l5;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
|
| 13 | d2uubk1 |
|
 |
97.4 |
21 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
| 14 | d2qalk1 |
|
 |
97.1 |
20 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
| 15 | c3bbnK_ |
|
 |
96.8 |
18 |
PDB header:ribosome
Chain: K: PDB Molecule:ribosomal protein s11;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 16 | c2zkqk_ |
|
 |
96.5 |
16 |
PDB header:ribosomal protein/rna
Chain: K: PDB Molecule:
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 17 | c3jyvK_ |
|
 |
95.1 |
20 |
PDB header:ribosome
Chain: K: PDB Molecule:40s ribosomal protein s14(a);
PDBTitle: structure of the 40s rrna and proteins and p/e trna for eukaryotic2 ribosome based on cryo-em map of thermomyces lanuginosus ribosome at3 8.9a resolution
|
| 18 | c2yfnA_ |
|
 |
70.6 |
31 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase-sucrose kinase agask;
PDBTitle: galactosidase domain of alpha-galactosidase-sucrose kinase,2 agask
|
| 19 | c3mi6A_ |
|
 |
63.9 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: crystal structure of the alpha-galactosidase from lactobacillus2 brevis, northeast structural genomics consortium target lbr11.
|
| 20 | c2xn1B_ |
|
 |
61.1 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-galactosidase;
PDBTitle: structure of alpha-galactosidase from lactobacillus acidophilus ncfm2 with tris
|
| 21 | c3gzaB_ |
|
not modelled |
59.5 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of putative alpha-l-fucosidase (np_812709.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.60 a resolution
|
| 22 | c2pohA_ |
|
not modelled |
50.6 |
50 |
PDB header:viral protein
Chain: A: PDB Molecule:head completion protein;
PDBTitle: structure of phage p22 tail needle gp26
|
| 23 | c3tevA_ |
|
not modelled |
45.1 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyl hyrolase, family 3;
PDBTitle: the crystal structure of glycosyl hydrolase from deinococcus2 radiodurans r1
|
| 24 | d1hl9a2 |
|
not modelled |
44.1 |
38 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Putative alpha-L-fucosidase, catalytic domain |
| 25 | c1hl8B_ |
|
not modelled |
43.0 |
38 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of thermotoga maritima alpha-fucosidase
|
| 26 | c2wvsD_ |
|
not modelled |
41.1 |
29 |
PDB header:hydrolase
Chain: D: PDB Molecule:alpha-l-fucosidase;
PDBTitle: crystal structure of an alpha-l-fucosidase gh29 trapped2 covalent intermediate from bacteroides thetaiotaomicron in3 complex with 2-fluoro-fucosyl fluoride using an e288q4 mutant
|
| 27 | c3ff4A_ |
|
not modelled |
40.0 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein chu_1412
|
| 28 | d1gtka2 |
|
not modelled |
37.1 |
18 |
Fold:dsRBD-like
Superfamily:Porphobilinogen deaminase (hydroxymethylbilane synthase), C-terminal domain
Family:Porphobilinogen deaminase (hydroxymethylbilane synthase), C-terminal domain |
| 29 | c3mo4B_ |
|
not modelled |
35.6 |
40 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-1,3/4-fucosidase;
PDBTitle: the crystal structure of an alpha-(1-3,4)-fucosidase from2 bifidobacterium longum subsp. infantis atcc 15697
|
| 30 | c3m0zD_ |
|
not modelled |
31.6 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:putative aldolase;
PDBTitle: crystal structure of putative aldolase from klebsiella2 pneumoniae.
|
| 31 | d1owla2 |
|
not modelled |
29.8 |
18 |
Fold:Cryptochrome/photolyase, N-terminal domain
Superfamily:Cryptochrome/photolyase, N-terminal domain
Family:Cryptochrome/photolyase, N-terminal domain |
| 32 | d1np7a2 |
|
not modelled |
28.6 |
14 |
Fold:Cryptochrome/photolyase, N-terminal domain
Superfamily:Cryptochrome/photolyase, N-terminal domain
Family:Cryptochrome/photolyase, N-terminal domain |
| 33 | c3eypB_ |
|
not modelled |
28.0 |
40 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative alpha-l-fucosidase;
PDBTitle: crystal structure of putative alpha-l-fucosidase from bacteroides2 thetaiotaomicron
|
| 34 | c3noyA_ |
|
not modelled |
27.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
| 35 | d3cuma2 |
|
not modelled |
25.0 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 36 | d1t35a_ |
|
not modelled |
24.8 |
36 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
| 37 | c3qk7C_ |
|
not modelled |
23.4 |
28 |
PDB header:transcription regulator
Chain: C: PDB Molecule:transcriptional regulators;
PDBTitle: crystal structure of putative transcriptional regulator from yersinia2 pestis biovar microtus str. 91001
|
| 38 | c2juiA_ |
|
not modelled |
23.0 |
32 |
PDB header:toxin
Chain: A: PDB Molecule:plne;
PDBTitle: three-dimensional structure of the two peptides that2 constitute the two-peptide bacteriocin plantaracin ef
|
| 39 | d1zpda2 |
|
not modelled |
21.3 |
22 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase Pyr module |
| 40 | c3qhaB_ |
|
not modelled |
20.7 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of a putative oxidoreductase from mycobacterium2 avium 104
|
| 41 | d1u3da2 |
|
not modelled |
19.9 |
12 |
Fold:Cryptochrome/photolyase, N-terminal domain
Superfamily:Cryptochrome/photolyase, N-terminal domain
Family:Cryptochrome/photolyase, N-terminal domain |
| 42 | c2cukC_ |
|
not modelled |
18.1 |
32 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerate dehydrogenase/glyoxylate reductase;
PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
|
| 43 | c2uyyD_ |
|
not modelled |
18.1 |
16 |
PDB header:cytokine
Chain: D: PDB Molecule:n-pac protein;
PDBTitle: structure of the cytokine-like nuclear factor n-pac
|
| 44 | d1kjqa2 |
|
not modelled |
17.4 |
29 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
| 45 | c2iz1C_ |
|
not modelled |
16.9 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: 6pdh complexed with pex inhibitor synchrotron data
|
| 46 | d1pdaa2 |
|
not modelled |
16.7 |
29 |
Fold:dsRBD-like
Superfamily:Porphobilinogen deaminase (hydroxymethylbilane synthase), C-terminal domain
Family:Porphobilinogen deaminase (hydroxymethylbilane synthase), C-terminal domain |
| 47 | d2djia2 |
|
not modelled |
15.6 |
28 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Pyruvate oxidase and decarboxylase Pyr module |
| 48 | c1vpdA_ |
|
not modelled |
15.5 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tartronate semialdehyde reductase;
PDBTitle: x-ray crystal structure of tartronate semialdehyde reductase2 [salmonella typhimurium lt2]
|
| 49 | c2duwA_ |
|
not modelled |
15.5 |
36 |
PDB header:ligand binding protein
Chain: A: PDB Molecule:putative coa-binding protein;
PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
|
| 50 | c3gybB_ |
|
not modelled |
15.5 |
11 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulators (laci-family
PDBTitle: crystal structure of a laci-family transcriptional2 regulatory protein from corynebacterium glutamicum
|
| 51 | d1vpda2 |
|
not modelled |
15.2 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 52 | d1y81a1 |
|
not modelled |
15.0 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 53 | c3eywA_ |
|
not modelled |
15.0 |
25 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
| 54 | d1dosa_ |
|
not modelled |
14.1 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
| 55 | c3qi7A_ |
|
not modelled |
14.1 |
29 |
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of a putative transcriptional regulator2 (yp_001089212.1) from clostridium difficile 630 at 1.86 a resolution
|
| 56 | d1np3a2 |
|
not modelled |
14.0 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 57 | c2w4lC_ |
|
not modelled |
13.7 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:deoxycytidylate deaminase;
PDBTitle: human dcmp deaminase
|
| 58 | c2ypnA_ |
|
not modelled |
13.3 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:protein (hydroxymethylbilane synthase);
PDBTitle: hydroxymethylbilane synthase
|
| 59 | d1ktba2 |
|
not modelled |
13.2 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 60 | d1hg3a_ |
|
not modelled |
13.2 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 61 | c3toxG_ |
|
not modelled |
12.3 |
28 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase in complex with2 nad(p) from sinorhizobium meliloti 1021
|
| 62 | c2gf2B_ |
|
not modelled |
12.2 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-hydroxyisobutyrate dehydrogenase;
PDBTitle: crystal structure of human hydroxyisobutyrate dehydrogenase
|
| 63 | d1qyda_ |
|
not modelled |
12.1 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 64 | d1w0ma_ |
|
not modelled |
12.1 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
| 65 | c3muxB_ |
|
not modelled |
12.0 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:putative 4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: the crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase2 from bacillus anthracis to 1.45a
|
| 66 | c3m6yA_ |
|
not modelled |
11.6 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase;
PDBTitle: structure of 4-hydroxy-2-oxoglutarate aldolase from bacillus cereus at2 1.45 a resolution.
|
| 67 | d1aopa1 |
|
not modelled |
11.4 |
25 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 68 | c2yicC_ |
|
not modelled |
11.2 |
22 |
PDB header:lyase
Chain: C: PDB Molecule:2-oxoglutarate decarboxylase;
PDBTitle: crystal structure of the suca domain of mycobacterium smegmatis2 alpha-ketoglutarate decarboxylase (triclinic form)
|
| 69 | d1pjca1 |
|
not modelled |
11.2 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 70 | c1ozhD_ |
|
not modelled |
11.1 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:acetolactate synthase, catabolic;
PDBTitle: the crystal structure of klebsiella pneumoniae acetolactate2 synthase with enzyme-bound cofactor and with an unusual3 intermediate.
|
| 71 | c3l6dB_ |
|
not modelled |
10.8 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of putative oxidoreductase from pseudomonas putida2 kt2440
|
| 72 | d1ae1a_ |
|
not modelled |
10.5 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 73 | c3u5in_ |
|
not modelled |
10.5 |
44 |
PDB header:ribosome
Chain: N: PDB Molecule:60s ribosomal protein l15-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
|
| 74 | c3u5en_ |
|
not modelled |
10.5 |
44 |
PDB header:ribosome
Chain: N: PDB Molecule:60s ribosomal protein l15-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
|
| 75 | c1tezB_ |
|
not modelled |
10.5 |
19 |
PDB header:lyase/dna
Chain: B: PDB Molecule:deoxyribodipyrimidine photolyase;
PDBTitle: complex between dna and the dna photolyase from anacystis nidulans
|
| 76 | c1zy9A_ |
|
not modelled |
10.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-galactosidase;
PDBTitle: crystal structure of alpha-galactosidase (ec 3.2.1.22) (melibiase)2 (tm1192) from thermotoga maritima at 2.34 a resolution
|
| 77 | d1hjqa_ |
|
not modelled |
10.5 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 78 | d1vq2a_ |
|
not modelled |
10.4 |
26 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
| 79 | d1pgja2 |
|
not modelled |
10.3 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 80 | d2i9ua1 |
|
not modelled |
10.2 |
14 |
Fold:Composite domain of metallo-dependent hydrolases
Superfamily:Composite domain of metallo-dependent hydrolases
Family:SAH/MTA deaminase-like |
| 81 | d2naca1 |
|
not modelled |
9.9 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 82 | d1p5dx1 |
|
not modelled |
9.8 |
24 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
| 83 | c3bmxB_ |
|
not modelled |
9.8 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:uncharacterized lipoprotein ybbd;
PDBTitle: beta-n-hexosaminidase (ybbd) from bacillus subtilis
|
| 84 | c3lk6A_ |
|
not modelled |
9.7 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipoprotein ybbd;
PDBTitle: beta-n-hexosaminidase n318d mutant (ybbd_n318d) from bacillus subtilis
|
| 85 | d2d59a1 |
|
not modelled |
9.6 |
36 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 86 | c3egcF_ |
|
not modelled |
9.6 |
15 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:putative ribose operon repressor;
PDBTitle: crystal structure of a putative ribose operon repressor from2 burkholderia thailandensis
|
| 87 | c2rafC_ |
|
not modelled |
9.5 |
12 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative dinucleotide-binding oxidoreductase;
PDBTitle: crystal structure of putative dinucleotide-binding2 oxidoreductase (np_786167.1) from lactobacillus plantarum3 at 1.60 a resolution
|
| 88 | d1gdha1 |
|
not modelled |
9.5 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 89 | c3sjuA_ |
|
not modelled |
9.5 |
35 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:keto reductase;
PDBTitle: hedamycin polyketide ketoreductase bound to nadph
|
| 90 | d1vp8a_ |
|
not modelled |
9.4 |
41 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 91 | d1zj8a1 |
|
not modelled |
9.3 |
22 |
Fold:Ferredoxin-like
Superfamily:Nitrite/Sulfite reductase N-terminal domain-like
Family:Duplicated SiR/NiR-like domains 1 and 3 |
| 92 | d1ebda2 |
|
not modelled |
9.2 |
13 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
| 93 | d1leha1 |
|
not modelled |
9.2 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 94 | d3coxa1 |
|
not modelled |
9.1 |
22 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
| 95 | c2fpgA_ |
|
not modelled |
8.9 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:succinyl-coa ligase [gdp-forming] alpha-chain,
PDBTitle: crystal structure of pig gtp-specific succinyl-coa2 synthetase in complex with gdp
|
| 96 | c3q94B_ |
|
not modelled |
8.9 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:fructose-bisphosphate aldolase, class ii;
PDBTitle: the crystal structure of fructose 1,6-bisphosphate aldolase from2 bacillus anthracis str. 'ames ancestor'
|
| 97 | c1zpdA_ |
|
not modelled |
8.8 |
26 |
PDB header:alcohol fermentation
Chain: A: PDB Molecule:pyruvate decarboxylase;
PDBTitle: pyruvate decarboxylase from zymomonas mobilis
|
| 98 | c3uf0A_ |
|
not modelled |
8.8 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: crystal structure of a putative nad(p) dependent gluconate 5-2 dehydrogenase from beutenbergia cavernae(efi target efi-502044) with3 bound nadp (low occupancy)
|
| 99 | c3jvdA_ |
|
not modelled |
8.6 |
15 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulators;
PDBTitle: crystal structure of putative transcription regulation repressor (laci2 family) from corynebacterium glutamicum
|

