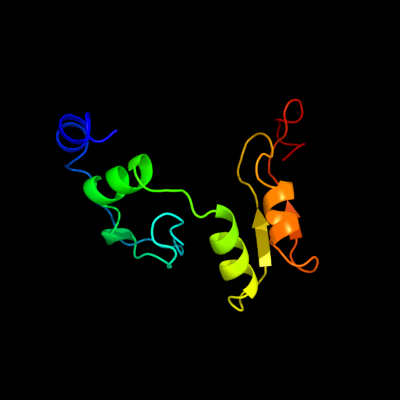
| 1 | c2qo3A_
|
|
 |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:eryaii erythromycin polyketide synthase modules 3 and 4;
PDBTitle: crystal structure of [ks3][at3] didomain from module 3 of 6-2 deoxyerthronolide b synthase
|
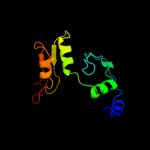
| 2 | c3eenA_
|
|
 |
99.9 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl coa-acp transacylase;
PDBTitle: crystal structure of malonyl-coa:acyl carrier protein transacylase2 (fabd), xoo0880, from xanthomonas oryzae pv. oryzae kacc10331
|
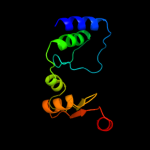
| 3 | d1nm2a1
|
|
 |
99.9 |
14 |
Fold:FabD/lysophospholipase-like
Superfamily:FabD/lysophospholipase-like
Family:FabD-like |
| 4 | c2qj3B_
|
|
 |
99.9 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:malonyl coa-acyl carrier protein transacylase;
PDBTitle: mycobacterium tuberculosis fabd
|
| 5 | c3im9A_
|
|
 |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl coa-acyl carrier protein transacylase;
PDBTitle: crystal structure of mcat from staphylococcus aureus
|
| 6 | c3ezoA_
|
|
 |
99.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl coa-acyl carrier protein transacylase;
PDBTitle: crystal structure of acyl-carrier-protein s-2 malonyltransferase from burkholderia pseudomallei 1710b
|
| 7 | d1mlaa1
|
|
 |
99.9 |
21 |
Fold:FabD/lysophospholipase-like
Superfamily:FabD/lysophospholipase-like
Family:FabD-like |
| 8 | c2hg4A_
|
|
 |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:6-deoxyerythronolide b synthase;
PDBTitle: structure of the ketosynthase-acyltransferase didomain of module 52 from debs.
|
| 9 | c2cuyA_
|
|
 |
99.9 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl coa-[acyl carrier protein] transacylase;
PDBTitle: crystal structure of malonyl coa-acyl carrier protein transacylase2 from thermus thermophilus hb8
|
| 10 | c3rgiA_
|
|
 |
99.9 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:disd protein;
PDBTitle: trans-acting transferase from disorazole synthase
|
| 11 | c2cdh9_
|
|
 |
99.9 |
16 |
PDB header:transferase
Chain: 9: PDB Molecule:acetyl transferase;
PDBTitle: architecture of the thermomyces lanuginosus fungal fatty2 acid synthase at 5 angstrom resolution.
|
| 12 | c3qatB_
|
|
 |
99.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:malonyl coa-acyl carrier protein transacylase;
PDBTitle: crystal structure of acyl-carrier-protein-s-malonyltransferase from2 bartonella henselae
|
| 13 | c2g2oA_
|
|
 |
99.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl coa-acyl carrier protein transacylase;
PDBTitle: structure of e.coli fabd complexed with sulfate
|
| 14 | c2c2nA_
|
|
 |
99.9 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl coa-acyl carrier protein transacylase;
PDBTitle: structure of human mitochondrial malonyltransferase
|
| 15 | c3ptwA_
|
|
 |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl coa-acyl carrier protein transacylase;
PDBTitle: crystal structure of malonyl coa-acyl carrier protein transacylase2 from clostridium perfringens atcc 13124
|
| 16 | c3tqeA_
|
|
 |
99.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl-coa-[acyl-carrier-protein] transacylase;
PDBTitle: structure of the malonyl coa-acyl carrier protein transacylase (fabd)2 from coxiella burnetii
|
| 17 | c2h1yA_
|
|
 |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl coenzyme a-acyl carrier protein transacylase;
PDBTitle: crystal structure of malonyl-coa:acyl carrier protein transacylase2 (mcat) from helicobacter pylori
|
| 18 | c3im8A_
|
|
 |
99.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl acyl carrier protein transacylase;
PDBTitle: crystal structure of mcat from streptococcus pneumoniae
|
| 19 | c3g87A_
|
|
 |
99.8 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:malonyl coa-acyl carrier protein transacylase;
PDBTitle: crystal structure of malonyl coa-acyl carrier protein transacylase2 from burkholderia pseudomallei using dried seaweed as nucleant or3 protease
|
| 20 | c2jfkD_
|
|
 |
99.8 |
17 |
PDB header:transferase
Chain: D: PDB Molecule:fatty acid synthase;
PDBTitle: structure of the mat domain of human fas with malonyl-coa
|
| 21 | c3hhdC_ |
|
not modelled |
99.8 |
17 |
PDB header:transferase, hydrolase
Chain: C: PDB Molecule:fatty acid synthase;
PDBTitle: structure of the human fatty acid synthase ks-mat didomain2 as a framework for inhibitor design.
|
| 22 | c2vz8B_ |
|
not modelled |
99.7 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase;
PDBTitle: crystal structure of mammalian fatty acid synthase
|
| 23 | c2uv9B_ |
|
not modelled |
99.0 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase alpha subunits;
PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the alpha subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
|
| 24 | c2uv8C_ |
|
not modelled |
98.7 |
27 |
PDB header:transferase
Chain: C: PDB Molecule:fatty acid synthase subunit alpha (fas2);
PDBTitle: crystal structure of yeast fatty acid synthase with stalled2 acyl carrier protein at 3.1 angstrom resolution
|
| 25 | c2vkzC_ |
|
not modelled |
98.7 |
27 |
PDB header:transferase
Chain: C: PDB Molecule:fatty acid synthase subunit alpha;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid2 synthase type i multienzyme complex
|
| 26 | c2vkzH_ |
|
not modelled |
97.8 |
15 |
PDB header:transferase
Chain: H: PDB Molecule:fatty acid synthase subunit beta;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid2 synthase type i multienzyme complex
|
| 27 | c2uvaI_ |
|
not modelled |
97.4 |
14 |
PDB header:transferase
Chain: I: PDB Molecule:fatty acid synthase beta subunits;
PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the beta subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
|
| 28 | c2xznC_ |
|
not modelled |
68.4 |
10 |
PDB header:ribosome
Chain: C: PDB Molecule:kh domain containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
| 29 | c2zkqc_ |
|
not modelled |
67.1 |
9 |
PDB header:ribosomal protein/rna
Chain: C: PDB Molecule:rna expansion segment es4;
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 30 | c1s1hC_ |
|
not modelled |
59.7 |
5 |
PDB header:ribosome
Chain: C: PDB Molecule:40s ribosomal protein s3;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
|
| 31 | c3izbB_ |
|
not modelled |
53.4 |
5 |
PDB header:ribosome
Chain: B: PDB Molecule:40s ribosomal protein rps3 (s3p);
PDBTitle: localization of the small subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
|
| 32 | c3iraA_ |
|
not modelled |
52.2 |
32 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein;
PDBTitle: the crystal structure of one domain of the conserved protein from2 methanosarcina mazei go1
|
| 33 | c3ea0B_ |
|
not modelled |
38.3 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:atpase, para family;
PDBTitle: crystal structure of para family atpase from chlorobium tepidum tls
|
| 34 | c3fhkF_ |
|
not modelled |
23.8 |
27 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:upf0403 protein yphp;
PDBTitle: crystal structure of apc1446, b.subtilis yphp disulfide2 isomerase
|
| 35 | c1x0uB_ |
|
not modelled |
13.7 |
10 |
PDB header:lyase
Chain: B: PDB Molecule:hypothetical methylmalonyl-coa decarboxylase alpha subunit;
PDBTitle: crystal structure of the carboxyl transferase subunit of putative pcc2 of sulfolobus tokodaii
|
| 36 | c3u9rB_ |
|
not modelled |
12.8 |
9 |
PDB header:ligase
Chain: B: PDB Molecule:methylcrotonyl-coa carboxylase, beta-subunit;
PDBTitle: crystal structure of p. aeruginosa 3-methylcrotonyl-coa carboxylase2 (mcc), beta subunit
|
| 37 | c2p4gA_ |
|
not modelled |
11.3 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a pyrimidine reductase-like protein (dip1392)2 from corynebacterium diphtheriae nctc at 2.30 a resolution
|
| 38 | d2qalc2 |
|
not modelled |
9.7 |
13 |
Fold:Ribosomal protein S3 C-terminal domain
Superfamily:Ribosomal protein S3 C-terminal domain
Family:Ribosomal protein S3 C-terminal domain |
| 39 | c1hnwC_ |
|
not modelled |
9.6 |
9 |
PDB header:ribosome
Chain: C: PDB Molecule:30s ribosomal protein s3;
PDBTitle: structure of the thermus thermophilus 30s ribosomal subunit2 in complex with tetracycline
|
| 40 | c1pnxC_ |
|
not modelled |
9.6 |
9 |
PDB header:ribosome
Chain: C: PDB Molecule:30s ribosomal protein s3;
PDBTitle: crystal structure of the wild type ribosome from e. coli,2 30s subunit of 70s ribosome. this file, 1pnx, contains3 only molecules of the 30s ribosomal subunit. the 50s4 subunit is in the pdb file 1pny.
|
| 41 | d2uubc2 |
|
not modelled |
8.7 |
0 |
Fold:Ribosomal protein S3 C-terminal domain
Superfamily:Ribosomal protein S3 C-terminal domain
Family:Ribosomal protein S3 C-terminal domain |
| 42 | c3n6rF_ |
|
not modelled |
8.7 |
10 |
PDB header:ligase
Chain: F: PDB Molecule:propionyl-coa carboxylase, beta subunit;
PDBTitle: crystal structure of the holoenzyme of propionyl-coa carboxylase (pcc)
|
| 43 | d1szaa_ |
|
not modelled |
8.2 |
13 |
Fold:alpha-alpha superhelix
Superfamily:ENTH/VHS domain
Family:RPR domain (SMART 00582 ) |
| 44 | d1oc4a2 |
|
not modelled |
7.3 |
10 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 45 | d1h05a_ |
|
not modelled |
7.3 |
46 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 46 | d1xb4a2 |
|
not modelled |
6.8 |
12 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Vacuolar sorting protein domain |
| 47 | c3uzuA_ |
|
not modelled |
6.7 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:ribosomal rna small subunit methyltransferase a;
PDBTitle: the structure of the ribosomal rna small subunit methyltransferase a2 from burkholderia pseudomallei
|
| 48 | c2gy9C_ |
|
not modelled |
6.6 |
13 |
PDB header:ribosome
Chain: C: PDB Molecule:30s ribosomal subunit protein s3;
PDBTitle: structure of the 30s subunit of a pre-translocational e.2 coli ribosome obtained by fitting atomic models for rna and3 protein components into cryo-em map emd-1056
|
| 49 | c2qbfC_ |
|
not modelled |
6.6 |
13 |
PDB header:ribosome
Chain: C: PDB Molecule:30s ribosomal protein s3;
PDBTitle: crystal structure of the bacterial ribosome from escherichia2 coli in complex with ribosome recycling factor (rrf). this3 file contains the 30s subunit of the second 70s ribosome.4 the entire crystal structure contains two 70s ribosomes and5 is described in remark 400.
|
| 50 | d1ez4a2 |
|
not modelled |
6.6 |
13 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 51 | d1zkda1 |
|
not modelled |
6.4 |
20 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:RPA4359-like |
| 52 | c1c4kA_ |
|
not modelled |
6.3 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:protein (ornithine decarboxylase);
PDBTitle: ornithine decarboxylase mutant (gly121tyr)
|
| 53 | c1i4wA_ |
|
not modelled |
6.2 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:mitochondrial replication protein mtf1;
PDBTitle: the crystal structure of the transcription factor sc-mttfb2 offers intriguing insights into mitochondrial transcription
|
| 54 | d1i4wa_ |
|
not modelled |
6.2 |
15 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:rRNA adenine dimethylase-like |
| 55 | c3n8kG_ |
|
not modelled |
6.1 |
44 |
PDB header:lyase
Chain: G: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: type ii dehydroquinase from mycobacterium tuberculosis complexed with2 citrazinic acid
|
| 56 | c2vycA_ |
|
not modelled |
5.8 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:biodegradative arginine decarboxylase;
PDBTitle: crystal structure of acid induced arginine decarboxylase2 from e. coli
|
| 57 | c2d5nB_ |
|
not modelled |
5.6 |
9 |
PDB header:hydrolase, oxidoreductase
Chain: B: PDB Molecule:riboflavin biosynthesis protein ribd;
PDBTitle: crystal structure of a bifunctional deaminase and reductase2 involved in riboflavin biosynthesis
|
| 58 | c3fteA_ |
|
not modelled |
5.6 |
12 |
PDB header:transferase/rna
Chain: A: PDB Molecule:dimethyladenosine transferase;
PDBTitle: crystal structure of a. aeolicus ksga in complex with rna
|
| 59 | d1o6za2 |
|
not modelled |
5.5 |
17 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
| 60 | c3d9oB_ |
|
not modelled |
5.5 |
11 |
PDB header:transcription
Chain: B: PDB Molecule:rna-binding protein 16;
PDBTitle: snapshots of the rna processing factor scaf8 bound to different2 phosphorylated forms of the carboxy-terminal domain of rna-polymerase3 ii
|
| 61 | c2uygF_ |
|
not modelled |
5.4 |
29 |
PDB header:lyase
Chain: F: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystallogaphic structure of the typeii 3-dehydroquinase2 from thermus thermophilus
|
| 62 | d2c4va1 |
|
not modelled |
5.4 |
29 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 63 | c2xigA_ |
|
not modelled |
5.0 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:ferric uptake regulation protein;
PDBTitle: the structure of the helicobacter pylori ferric uptake2 regulator fur reveals three functional metal binding sites
|