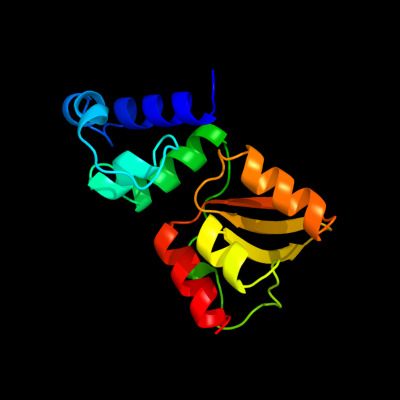
| 1 | d2d28c1
|
|
 |
100.0 |
18 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:EspE N-terminal domain-like
Family:GSPII protein E N-terminal domain-like |
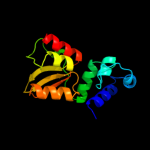
| 2 | c3f1yC_
|
|
 |
100.0 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: mannosyl-3-phosphoglycerate synthase from rubrobacter xylanophilus
|
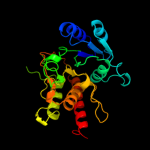
| 3 | c1xhbA_
|
|
 |
99.9 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 1;
PDBTitle: the crystal structure of udp-galnac: polypeptide alpha-n-2 acetylgalactosaminyltransferase-t1
|
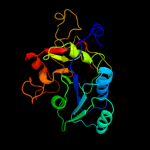
| 4 | d1xhba2
|
|
 |
99.9 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Polypeptide N-acetylgalactosaminyltransferase 1, N-terminal domain |
| 5 | c2z86D_
|
|
 |
99.9 |
20 |
PDB header:transferase
Chain: D: PDB Molecule:chondroitin synthase;
PDBTitle: crystal structure of chondroitin polymerase from2 escherichia coli strain k4 (k4cp) complexed with udp-glcua3 and udp
|
| 6 | c2d7iA_
|
|
 |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 10;
PDBTitle: crsytal structure of pp-galnac-t10 with udp, galnac and mn2+
|
| 7 | c2ffuA_
|
|
 |
99.9 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:polypeptide n-acetylgalactosaminyltransferase 2;
PDBTitle: crystal structure of human ppgalnact-2 complexed with udp2 and ea2
|
| 8 | c3ckvA_
|
|
 |
99.9 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a mycobacterial protein
|
| 9 | c3bcvA_
|
|
 |
99.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:putative glycosyltransferase protein;
PDBTitle: crystal structure of a putative glycosyltransferase from bacteroides2 fragilis
|
| 10 | d1qg8a_
|
|
 |
99.9 |
8 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Spore coat polysaccharide biosynthesis protein SpsA |
| 11 | c1omxB_
|
|
 |
99.8 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:alpha-1,4-n-acetylhexosaminyltransferase extl2;
PDBTitle: crystal structure of mouse alpha-1,4-n-2 acetylhexosaminyltransferase (extl2)
|
| 12 | d1omza_
|
|
 |
99.8 |
12 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Exostosin |
| 13 | c2qgiA_
|
|
 |
99.6 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:atp synthase subunits region orf 6;
PDBTitle: the udp complex structure of the sixth gene product of the f1-atpase2 operon of rhodobacter blasticus
|
| 14 | d2bo4a1
|
|
 |
99.3 |
17 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:MGS-like |
| 15 | d1pzta_
|
|
 |
98.6 |
18 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:beta 1,4 galactosyltransferase (b4GalT1) |
| 16 | c3lw6A_
|
|
 |
98.2 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:beta-4-galactosyltransferase 7;
PDBTitle: crystal structure of drosophila beta1,4-galactosyltransferase-7
|
| 17 | d1fo8a_
|
|
 |
97.8 |
8 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:N-acetylglucosaminyltransferase I |
| 18 | d1p9ra_
|
|
 |
97.0 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 19 | c2wvmA_
|
|
 |
95.1 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: h309a mutant of mannosyl-3-phosphoglycerate synthase from2 thermus thermophilus hb27 in complex with3 gdp-alpha-d-mannose and mg(ii)
|
| 20 | c2zu8A_
|
|
 |
94.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:mannosyl-3-phosphoglycerate synthase;
PDBTitle: crystal structure of mannosyl-3-phosphoglycerate synthase2 from pyrococcus horikoshii
|
| 21 | c2bh1Y_ |
|
not modelled |
94.0 |
15 |
PDB header:transport protein
Chain: Y: PDB Molecule:general secretion pathway protein e,;
PDBTitle: x-ray structure of the general secretion pathway complex of2 the n-terminal domain of epse and the cytosolic domain of3 epsl of vibrio cholerae
|
| 22 | d2bh1x1 |
|
not modelled |
94.0 |
15 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:EspE N-terminal domain-like
Family:GSPII protein E N-terminal domain-like |
| 23 | c2ch4A_
|
|
 |
89.1 |
10 |
PDB header:transferase/chemotaxis
Chain: A: PDB Molecule:chemotaxis protein chea;
PDBTitle: complex between bacterial chemotaxis histidine kinase chea2 domains p4 and p5 and receptor-adaptor protein chew
|
| 24 | c2oaq1_ |
|
not modelled |
83.3 |
7 |
PDB header:hydrolase
Chain: 1: PDB Molecule:type ii secretion system protein;
PDBTitle: crystal structure of the archaeal secretion atpase gspe in complex2 with phosphate
|
| 25 | d1ugpa_
|
|
 |
68.3 |
18 |
Fold:Nitrile hydratase alpha chain
Superfamily:Nitrile hydratase alpha chain
Family:Nitrile hydratase alpha chain |
| 26 | c1jylC_ |
|
not modelled |
65.6 |
14 |
PDB header:transferase
Chain: C: PDB Molecule:ctp:phosphocholine cytidylytransferase;
PDBTitle: catalytic mechanism of ctp:phosphocholine2 cytidylytransferase from streptococcus pneumoniae (licc)
|
| 27 | c3qyhG_ |
|
not modelled |
63.9 |
19 |
PDB header:lyase
Chain: G: PDB Molecule:co-type nitrile hydratase alpha subunit;
PDBTitle: crystal structure of co-type nitrile hydratase beta-h71l from2 pseudomonas putida.
|
| 28 | c2dd4H_ |
|
not modelled |
58.1 |
14 |
PDB header:hydrolase
Chain: H: PDB Molecule:thiocyanate hydrolase beta subunit;
PDBTitle: thiocyanate hydrolase (scnase) from thiobacillus thioparus2 recombinant apo-enzyme
|
| 29 | d1ugpb_ |
|
not modelled |
56.3 |
11 |
Fold:SH3-like barrel
Superfamily:Electron transport accessory proteins
Family:Nitrile hydratase beta chain |
| 30 | c2gamA_ |
|
not modelled |
56.2 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:beta-1,6-n-acetylglucosaminyltransferase;
PDBTitle: x-ray crystal structure of murine leukocyte-type core 2 b1,2 6-n-acetylglucosaminyltransferase (c2gnt-l) in complex3 with galb1,3galnac
|
| 31 | c2qh5B_ |
|
not modelled |
46.6 |
10 |
PDB header:isomerase
Chain: B: PDB Molecule:mannose-6-phosphate isomerase;
PDBTitle: crystal structure of mannose-6-phosphate isomerase from helicobacter2 pylori
|
| 32 | c2j0bA_ |
|
not modelled |
44.9 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:beta-1,3-n-acetylglucosaminyltransferase manic fringe;
PDBTitle: structure of the catalytic domain of mouse manic fringe in2 complex with udp and manganese
|
| 33 | d1v29b_ |
|
not modelled |
43.6 |
10 |
Fold:SH3-like barrel
Superfamily:Electron transport accessory proteins
Family:Nitrile hydratase beta chain |
| 34 | d1v29a_ |
|
not modelled |
42.2 |
14 |
Fold:Nitrile hydratase alpha chain
Superfamily:Nitrile hydratase alpha chain
Family:Nitrile hydratase alpha chain |
| 35 | c2dxbR_ |
|
not modelled |
39.3 |
8 |
PDB header:hydrolase
Chain: R: PDB Molecule:thiocyanate hydrolase subunit gamma;
PDBTitle: recombinant thiocyanate hydrolase comprising partially-modified cobalt2 centers
|
| 36 | c2xmhB_ |
|
not modelled |
39.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:ctp-inositol-1-phosphate cytidylyltransferase;
PDBTitle: the x-ray structure of ctp:inositol-1-phosphate2 cytidylyltransferase from archaeoglobus fulgidus
|
| 37 | c2xzmD_ |
|
not modelled |
37.0 |
16 |
PDB header:ribosome
Chain: D: PDB Molecule:ribosomal protein s4 containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
|
| 38 | d2qdyb1 |
|
not modelled |
36.3 |
14 |
Fold:SH3-like barrel
Superfamily:Electron transport accessory proteins
Family:Nitrile hydratase beta chain |
| 39 | d1fxoa_ |
|
not modelled |
35.6 |
10 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 40 | d1jyka_ |
|
not modelled |
34.8 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 41 | c3kvhA_ |
|
not modelled |
30.9 |
22 |
PDB header:rna binding protein
Chain: A: PDB Molecule:protein syndesmos;
PDBTitle: crystal structure of human protein syndesmos (nudt16-like protein)
|
| 42 | d1fuoa_ |
|
not modelled |
30.0 |
17 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 43 | c1w57A_ |
|
not modelled |
30.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:ispd/ispf bifunctional enzyme;
PDBTitle: structure of the bifunctional ispdf from campylobacter2 jejuni containing zn
|
| 44 | c3jvvA_ |
|
not modelled |
28.7 |
9 |
PDB header:atp binding protein
Chain: A: PDB Molecule:twitching mobility protein;
PDBTitle: crystal structure of p. aeruginosa pilt with bound amp-pcp
|
| 45 | d1vh3a_ |
|
not modelled |
28.4 |
11 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
| 46 | d1v82a_ |
|
not modelled |
27.1 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:1,3-glucuronyltransferase |
| 47 | d1kyqa2 |
|
not modelled |
26.6 |
18 |
Fold:Siroheme synthase middle domains-like
Superfamily:Siroheme synthase middle domains-like
Family:Siroheme synthase middle domains-like |
| 48 | c3brkX_ |
|
not modelled |
26.1 |
13 |
PDB header:transferase
Chain: X: PDB Molecule:glucose-1-phosphate adenylyltransferase;
PDBTitle: crystal structure of adp-glucose pyrophosphorylase from2 agrobacterium tumefaciens
|
| 49 | d1lm5a_ |
|
not modelled |
25.4 |
10 |
Fold:beta-hairpin-alpha-hairpin repeat
Superfamily:Plakin repeat
Family:Plakin repeat |
| 50 | d2p12a1 |
|
not modelled |
25.0 |
14 |
Fold:FomD barrel-like
Superfamily:FomD-like
Family:FomD-like |
| 51 | c3fesB_ |
|
not modelled |
24.1 |
10 |
PDB header:atp binding protein
Chain: B: PDB Molecule:atp-dependent clp endopeptidase;
PDBTitle: crystal structure of the atp-dependent clp protease clpc from2 clostridium difficile
|
| 52 | c2x5sB_ |
|
not modelled |
23.6 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:mannose-1-phosphate guanylyltransferase;
PDBTitle: crystal structure of t. maritima gdp-mannose2 pyrophosphorylase in apo state.
|
| 53 | d1lvwa_ |
|
not modelled |
23.2 |
16 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 54 | d1jswa_ |
|
not modelled |
23.2 |
24 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 55 | d1j3ua_ |
|
not modelled |
21.7 |
23 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 56 | d2qdya1 |
|
not modelled |
21.6 |
11 |
Fold:Nitrile hydratase alpha chain
Superfamily:Nitrile hydratase alpha chain
Family:Nitrile hydratase alpha chain |
| 57 | d2oqoa1 |
|
not modelled |
21.3 |
22 |
Fold:Lysozyme-like
Superfamily:Lysozyme-like
Family:PBP transglycosylase domain-like |
| 58 | c2d0jD_ |
|
not modelled |
20.7 |
14 |
PDB header:transferase
Chain: D: PDB Molecule:galactosylgalactosylxylosylprotein 3-beta-
PDBTitle: crystal structure of human glcat-s apo form
|
| 59 | c3mwmA_ |
|
not modelled |
19.0 |
20 |
PDB header:transcription
Chain: A: PDB Molecule:putative metal uptake regulation protein;
PDBTitle: graded expression of zinc-responsive genes through two regulatory2 zinc-binding sites in zur
|
| 60 | c1chmA_ |
|
not modelled |
19.0 |
21 |
PDB header:creatinase
Chain: A: PDB Molecule:creatine amidinohydrolase;
PDBTitle: enzymatic mechanism of creatine amidinohydrolase as deduced2 from crystal structures
|
| 61 | d1zq1c3 |
|
not modelled |
16.8 |
15 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:GatB/GatE catalytic domain-like |
| 62 | d1dgna_ |
|
not modelled |
15.8 |
22 |
Fold:DEATH domain
Superfamily:DEATH domain
Family:Caspase recruitment domain, CARD |
| 63 | c2nz7A_ |
|
not modelled |
15.7 |
15 |
PDB header:apoptosis
Chain: A: PDB Molecule:caspase recruitment domain-containing protein 4;
PDBTitle: crystal structure analysis of caspase-recruitment domain2 (card) of nod1
|
| 64 | d2jera1 |
|
not modelled |
15.7 |
16 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Porphyromonas-type peptidylarginine deiminase |
| 65 | c2jerG_ |
|
not modelled |
15.3 |
16 |
PDB header:hydrolase
Chain: G: PDB Molecule:agmatine deiminase;
PDBTitle: agmatine deiminase of enterococcus faecalis catalyzing its2 reaction.
|
| 66 | c3bjrA_ |
|
not modelled |
15.2 |
5 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative carboxylesterase;
PDBTitle: crystal structure of a putative carboxylesterase (lp_1002) from2 lactobacillus plantarum wcfs1 at 2.09 a resolution
|
| 67 | d1yp2a2 |
|
not modelled |
15.0 |
13 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 68 | c1hsjA_ |
|
not modelled |
15.0 |
13 |
PDB header:transcription/sugar binding protein
Chain: A: PDB Molecule:fusion protein consisting of staphylococcus
PDBTitle: sarr mbp fusion structure
|
| 69 | d1z05a1 |
|
not modelled |
14.8 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:ROK associated domain |
| 70 | c1yfmA_ |
|
not modelled |
14.7 |
28 |
PDB header:lyase
Chain: A: PDB Molecule:fumarase;
PDBTitle: recombinant yeast fumarase
|
| 71 | d1yfma_ |
|
not modelled |
14.7 |
28 |
Fold:L-aspartase-like
Superfamily:L-aspartase-like
Family:L-aspartase/fumarase |
| 72 | c3qz9D_ |
|
not modelled |
14.4 |
20 |
PDB header:lyase
Chain: D: PDB Molecule:co-type nitrile hydratase beta subunit;
PDBTitle: crystal structure of co-type nitrile hydratase beta-y215f from2 pseudomonas putida.
|
| 73 | d1u20a1 |
|
not modelled |
14.0 |
18 |
Fold:Nudix
Superfamily:Nudix
Family:MutT-like |
| 74 | c1egpA_ |
|
not modelled |
13.8 |
24 |
PDB header:proteinase inhibitor
Chain: A: PDB Molecule:eglin-c;
PDBTitle: proteinase inhibitor eglin c with hydrolysed reactive center
|
| 75 | d2ewoa1 |
|
not modelled |
13.7 |
9 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Porphyromonas-type peptidylarginine deiminase |
| 76 | c3hzsA_ |
|
not modelled |
13.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:monofunctional glycosyltransferase;
PDBTitle: s. aureus monofunctional glycosyltransferase (mtga)in complex with2 moenomycin
|
| 77 | d1xkna_ |
|
not modelled |
13.3 |
9 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Porphyromonas-type peptidylarginine deiminase |
| 78 | c1ex1A_ |
|
not modelled |
13.3 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein (beta-d-glucan exohydrolase isoenzyme exo1);
PDBTitle: beta-d-glucan exohydrolase from barley
|
| 79 | d3crda_ |
|
not modelled |
12.8 |
15 |
Fold:DEATH domain
Superfamily:DEATH domain
Family:Caspase recruitment domain, CARD |
| 80 | d2f2ab2 |
|
not modelled |
12.4 |
19 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:GatB/GatE catalytic domain-like |
| 81 | c2jciA_ |
|
not modelled |
12.4 |
9 |
PDB header:drug-binding protein
Chain: A: PDB Molecule:penicillin-binding protein 1b;
PDBTitle: structural insights into the catalytic mechanism and the2 role of streptococcus pneumoniae pbp1b
|
| 82 | d1zbra1 |
|
not modelled |
12.1 |
16 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Porphyromonas-type peptidylarginine deiminase |
| 83 | c2wawA_ |
|
not modelled |
11.9 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:moba relate protein;
PDBTitle: crystal structure of mycobacterium tuberculosis rv0371c2 homolog from mycobacterium sp. strain jc1
|
| 84 | d2h80a1 |
|
not modelled |
11.6 |
29 |
Fold:SAM domain-like
Superfamily:SAM/Pointed domain
Family:Variant SAM domain |
| 85 | c1s1hO_ |
|
not modelled |
11.5 |
18 |
PDB header:ribosome
Chain: O: PDB Molecule:40s ribosomal protein s13;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
|
| 86 | d2oi6a2 |
|
not modelled |
11.0 |
8 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:UDP-glucose pyrophosphorylase |
| 87 | d1iina_ |
|
not modelled |
10.9 |
11 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 88 | c2xnsC_ |
|
not modelled |
10.8 |
21 |
PDB header:hydrolase/peptide
Chain: C: PDB Molecule:regulator of g-protein signaling 14;
PDBTitle: crystal structure of human g alpha i1 bound to a designed helical2 peptide derived from the goloco motif of rgs14
|
| 89 | c2d7cD_ |
|
not modelled |
10.0 |
25 |
PDB header:protein transport
Chain: D: PDB Molecule:rab11 family-interacting protein 3;
PDBTitle: crystal structure of human rab11 in complex with fip3 rab-2 binding domain
|
| 90 | d2pnwa1 |
|
not modelled |
9.7 |
24 |
Fold:Double psi beta-barrel
Superfamily:Barwin-like endoglucanases
Family:MLTA-like |
| 91 | c2gszE_ |
|
not modelled |
9.2 |
13 |
PDB header:protein transport
Chain: E: PDB Molecule:twitching motility protein pilt;
PDBTitle: structure of a. aeolicus pilt with 6 monomers per2 asymmetric unit
|
| 92 | d1mc3a_ |
|
not modelled |
9.0 |
10 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:glucose-1-phosphate thymidylyltransferase |
| 93 | c2o03A_ |
|
not modelled |
8.7 |
28 |
PDB header:gene regulation
Chain: A: PDB Molecule:probable zinc uptake regulation protein furb;
PDBTitle: crystal structure of furb from m. tuberculosis- a zinc uptake2 regulator
|
| 94 | d1mzba_ |
|
not modelled |
8.7 |
26 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:FUR-like |
| 95 | d1g5ta_ |
|
not modelled |
8.5 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
| 96 | c3udiA_ |
|
not modelled |
8.5 |
17 |
PDB header:penicillin-binding protein/antibiotic
Chain: A: PDB Molecule:penicillin-binding protein 1a;
PDBTitle: crystal structure of acinetobacter baumannii pbp1a in complex with2 penicillin g
|
| 97 | d1vkpa_ |
|
not modelled |
8.5 |
6 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Porphyromonas-type peptidylarginine deiminase |
| 98 | c3hxkB_ |
|
not modelled |
8.5 |
12 |
PDB header:hydrolase
Chain: B: PDB Molecule:sugar hydrolase;
PDBTitle: crystal structure of a sugar hydrolase (yeeb) from2 lactococcus lactis, northeast structural genomics3 consortium target kr108
|
| 99 | c1yshE_ |
|
not modelled |
8.4 |
18 |
PDB header:structural protein/rna
Chain: E: PDB Molecule:40s ribosomal protein s13;
PDBTitle: localization and dynamic behavior of ribosomal protein l30e
|