1 d1f7ua1
99.4
20
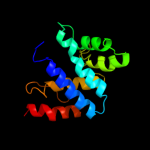
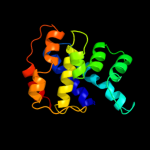
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases2 c1f7uA_
98.4
20
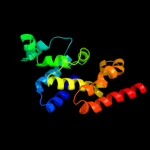
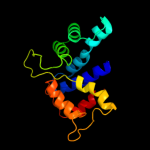
PDB header: ligase/rnaChain: A: PDB Molecule: arginyl-trna synthetase;PDBTitle: crystal structure of the arginyl-trna synthetase complexed with the2 trna(arg) and l-arg
3 c2zufA_
98.0
17
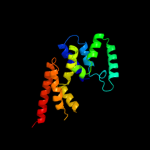
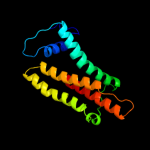
PDB header: ligase/rnaChain: A: PDB Molecule: arginyl-trna synthetase;PDBTitle: crystal structure of pyrococcus horikoshii arginyl-trna2 synthetase complexed with trna(arg)
4 c1iq0A_
97.9
20
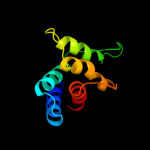
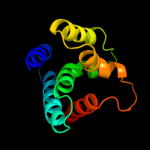
PDB header: ligaseChain: A: PDB Molecule: arginyl-trna synthetase;PDBTitle: thermus thermophilus arginyl-trna synthetase
5 c3gw7A_
97.9
19
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein yedj;PDBTitle: crystal structure of a metal-dependent phosphohydrolase2 with conserved hd domain (yedj) from escherichia coli in3 complex with nickel ions. northeast structural genomics4 consortium target er63
6 d1iq0a1
97.6
19
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases7 c2o08B_
97.5
23
PDB header: hydrolaseChain: B: PDB Molecule: bh1327 protein;PDBTitle: crystal structure of a putative hd superfamily hydrolase (bh1327) from2 bacillus halodurans at 1.90 a resolution
8 d2pq7a1
97.3
20
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain9 d2pjqa1
97.3
16
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain10 d2qgsa1
97.2
14
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain11 c3ccgA_
97.2
20
PDB header: hydrolaseChain: A: PDB Molecule: hd superfamily hydrolase;PDBTitle: crystal structure of predicted hd superfamily hydrolase involved in2 nad metabolism (np_347894.1) from clostridium acetobutylicum at 1.503 a resolution
12 c3fnrA_
97.2
14
PDB header: transferaseChain: A: PDB Molecule: arginyl-trna synthetase;PDBTitle: crystal structure of putative arginyl t-rna synthetase from2 campylobacter jejuni;
13 c2ogiA_
96.8
20
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein sag1661;PDBTitle: crystal structure of a putative metal dependent phosphohydrolase2 (sag1661) from streptococcus agalactiae serogroup v at 1.85 a3 resolution
14 d1r89a3
96.7
12
Fold: Ferredoxin-likeSuperfamily: PAP/Archaeal CCA-adding enzyme, C-terminal domainFamily: Archaeal tRNA CCA-adding enzyme15 d3djba1
96.3
16
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain16 c3hc1A_
95.9
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized hdod domain protein;PDBTitle: crystal structure of hdod domain protein with unknown function2 (np_953345.1) from geobacter sulfurreducens at 1.90 a resolution
17 c3m1tA_
95.8
19
PDB header: hydrolaseChain: A: PDB Molecule: putative phosphohydrolase;PDBTitle: crystal structure of putative phosphohydrolase (yp_929327.1) from2 shewanella amazonensis sb2b at 1.62 a resolution
18 c2x1lC_
95.6
16
PDB header: ligaseChain: C: PDB Molecule: methionyl-trna synthetase;PDBTitle: crystal structure of mycobacterium smegmatis methionyl-trna2 synthetase in complex with methionine and adenosine
19 d3dtoa1
95.6
17
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain20 c1sz1A_
94.4
13
PDB header: transferase/rnaChain: A: PDB Molecule: trna nucleotidyltransferase;PDBTitle: mechanism of cca-adding enzymes specificity revealed by crystal2 structures of ternary complexes
21 c3kflA_
not modelled
94.3
13
PDB header: ligaseChain: A: PDB Molecule: methionyl-trna synthetase;PDBTitle: leishmania major methionyl-trna synthetase in complex with2 methionyladenylate and pyrophosphate
22 d3b57a1
not modelled
94.2
13
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain23 c1pfuA_
not modelled
93.6
13
PDB header: ligaseChain: A: PDB Molecule: methionyl-trna synthetase;PDBTitle: methionyl-trna synthetase from escherichia coli complexed2 with methionine phosphinate
24 c2ct8A_
not modelled
92.3
18
PDB header: ligase/rnaChain: A: PDB Molecule: methionyl-trna synthetase;PDBTitle: crystal structure of aquifex aeolicus methionyl-trna2 synthetase complexed with trna(met) and methionyl-adenylate3 anologue
25 c1rqgA_
not modelled
91.2
11
PDB header: ligaseChain: A: PDB Molecule: methionyl-trna synthetase;PDBTitle: methionyl-trna synthetase from pyrococcus abyssi
26 c1woyA_
not modelled
88.8
17
PDB header: ligaseChain: A: PDB Molecule: methionyl-trna synthetase;PDBTitle: crystal structure of methionyl trna synthetase y225f mutant2 from thermus thermophilus
27 c2ztgA_
not modelled
87.8
21
PDB header: ligaseChain: A: PDB Molecule: alanyl-trna synthetase;PDBTitle: crystal structure of archaeoglobus fulgidus alanyl-trna2 synthetase lacking the c-terminal dimerization domain in3 complex with ala-sa
28 d1ynba1
not modelled
87.6
17
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain29 c2cqzA_
not modelled
85.2
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 177aa long hypothetical protein;PDBTitle: crystal structure of ph0347 protein from pyrococcus horikoshii ot3
30 c3i7aA_
not modelled
82.0
15
PDB header: hydrolaseChain: A: PDB Molecule: putative metal-dependent phosphohydrolase;PDBTitle: crystal structure of putative metal-dependent phosphohydrolase2 (yp_926882.1) from shewanella amazonensis sb2b at 2.06 a resolution
31 d1pfva1
not modelled
79.0
12
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases32 c2zzfA_
not modelled
73.5
17
PDB header: ligaseChain: A: PDB Molecule: alanyl-trna synthetase;PDBTitle: crystal structure of alanyl-trna synthetase without2 oligomerization domain
33 d2paqa1
not modelled
72.1
9
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain34 d1rqga1
not modelled
67.7
11
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases35 d1vqra_
not modelled
67.4
15
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: modified HD domain36 c3d5lA_
not modelled
63.9
5
PDB header: signaling proteinChain: A: PDB Molecule: regulatory protein recx;PDBTitle: crystal structure of regulatory protein recx
37 d1xx7a_
not modelled
63.0
17
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain38 d1riqa1
not modelled
54.4
15
Fold: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)Superfamily: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)Family: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)39 c3mzoA_
not modelled
51.7
16
PDB header: hydrolaseChain: A: PDB Molecule: lin2634 protein;PDBTitle: crystal structure of a hd-domain phosphohydrolase (lin2634) from2 listeria innocua at 1.98 a resolution
40 c3sp1B_
not modelled
48.2
19
PDB header: ligaseChain: B: PDB Molecule: cysteinyl-trna synthetase;PDBTitle: crystal structure of cysteinyl-trna synthetase (cyss) from borrelia2 burgdorferi
41 c1yfsB_
not modelled
45.0
15
PDB header: ligaseChain: B: PDB Molecule: alanyl-trna synthetase;PDBTitle: the crystal structure of alanyl-trna synthetase in complex2 with l-alanine
42 c2k89A_
not modelled
38.4
10
PDB header: protein bindingChain: A: PDB Molecule: phospholipase a-2-activating protein;PDBTitle: solution structure of a novel ubiquitin-binding domain from2 human plaa (pfuc, gly76-pro77 cis isomer)
43 c1sb7A_
not modelled
38.4
10
PDB header: lyaseChain: A: PDB Molecule: trna pseudouridine synthase d;PDBTitle: crystal structure of the e.coli pseudouridine synthase trud
44 c1z2zB_
not modelled
38.0
9
PDB header: lyaseChain: B: PDB Molecule: probable trna pseudouridine synthase d;PDBTitle: crystal structure of the putative trna pseudouridine2 synthase d (trud) from methanosarcina mazei, northeast3 structural genomics target mar1
45 c2gfnA_
not modelled
37.9
21
PDB header: transcriptionChain: A: PDB Molecule: hth-type transcriptional regulator pksa related protein;PDBTitle: crystal structure of hth-type transcriptional regulator pksa related2 protein from rhodococcus sp. rha1
46 d1pkxa1
not modelled
36.7
27
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Inosicase47 c3kh1B_
not modelled
36.0
12
PDB header: hydrolaseChain: B: PDB Molecule: predicted metal-dependent phosphohydrolase;PDBTitle: crystal structure of predicted metal-dependent2 phosphohydrolase (zp_00055740.2) from magnetospirillum3 magnetotacticum ms-1 at 1.37 a resolution
48 d2d5ba1
not modelled
35.5
17
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases49 c1hynQ_
not modelled
34.0
15
PDB header: membrane proteinChain: Q: PDB Molecule: band 3 anion transport protein;PDBTitle: crystal structure of the cytoplasmic domain of human2 erythrocyte band-3 protein
50 c3l3fX_
not modelled
33.5
19
PDB header: protein bindingChain: X: PDB Molecule: protein doa1;PDBTitle: crystal structure of a pfu-pul domain pair of saccharomyces cerevisiae2 doa1/ufd3
51 c3ljvA_
not modelled
33.3
21
PDB header: transcriptionChain: A: PDB Molecule: mmoq response regulator;PDBTitle: crystal structure of mmoq response regulator (fragment 29-302) from2 methylococcus capsulatus str. bath, northeast structural genomics3 consortium target mcr175m
52 c3dfgA_
not modelled
31.0
11
PDB header: recombinationChain: A: PDB Molecule: regulatory protein recx;PDBTitle: crystal structure of recx: a potent inhibitor protein of2 reca from xanthomonas campestris
53 d2gfna1
not modelled
30.1
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Tetracyclin repressor-like, N-terminal domain54 c3hxxA_
not modelled
28.8
17
PDB header: ligaseChain: A: PDB Molecule: alanyl-trna synthetase;PDBTitle: crystal structure of catalytic fragment of e. coli alars in complex2 with amppcp
55 c1ileA_
not modelled
27.3
10
PDB header: aminoacyl-trna synthetaseChain: A: PDB Molecule: isoleucyl-trna synthetase;PDBTitle: isoleucyl-trna synthetase
56 c3pstA_
not modelled
26.4
19
PDB header: nuclear proteinChain: A: PDB Molecule: protein doa1;PDBTitle: crystal structure of pul and pfu(mutate) domain
57 c3b81A_
not modelled
25.4
4
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, acrr family;PDBTitle: crystal structure of predicted dna-binding transcriptional regulator2 of tetr/acrr family (np_350189.1) from clostridium acetobutylicum at3 2.10 a resolution
58 c2zihC_
not modelled
23.9
16
PDB header: protein transportChain: C: PDB Molecule: vacuolar protein sorting-associated protein 74;PDBTitle: crystal structure of yeast vps74
59 c2o59B_
not modelled
23.9
16
PDB header: isomerase/dnaChain: B: PDB Molecule: dna topoisomerase 3;PDBTitle: structure of e. coli topoisomerase iii in complex with an 8-2 base single stranded oligonucleotide. frozen in glycerol3 ph 8.0
60 d1szwa_
not modelled
23.4
13
Fold: Pseudouridine synthaseSuperfamily: Pseudouridine synthaseFamily: tRNA pseudouridine synthase TruD61 c2ziiA_
not modelled
23.2
16
PDB header: protein transportChain: A: PDB Molecule: vacuolar protein sorting-associated protein 74;PDBTitle: crystal structure of yeast vps74-n-term truncation variant
62 d1hynp_
not modelled
22.9
17
Fold: Phoshotransferase/anion transport proteinSuperfamily: Phoshotransferase/anion transport proteinFamily: Anion transport protein, cytoplasmic domain63 d2guma1
not modelled
22.5
10
Fold: Viral glycoprotein ectodomain-likeSuperfamily: Viral glycoprotein ectodomain-likeFamily: Glycoprotein B-like64 c3kpbA_
not modelled
22.2
12
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein mj0100;PDBTitle: crystal structure of the cbs domain pair of protein mj01002 in complex with 5 -methylthioadenosine and s-adenosyl-l-3 methionine.
65 c3orgB_
not modelled
21.0
31
PDB header: transport proteinChain: B: PDB Molecule: cmclc;PDBTitle: crystal structure of a eukaryotic clc transporter
66 c2qv5A_
not modelled
20.9
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu2773;PDBTitle: crystal structure of uncharacterized protein atu2773 from2 agrobacterium tumefaciens c58
67 c3nw8B_
not modelled
20.4
10
PDB header: viral proteinChain: B: PDB Molecule: envelope glycoprotein b;PDBTitle: glycoprotein b from herpes simplex virus type 1, y179s mutant, high-ph
68 c3e3vA_
not modelled
19.9
21
PDB header: recombinationChain: A: PDB Molecule: regulatory protein recx;PDBTitle: crystal structure of recx from lactobacillus salivarius
69 d1i7da_
not modelled
19.4
19
Fold: Prokaryotic type I DNA topoisomeraseSuperfamily: Prokaryotic type I DNA topoisomeraseFamily: Prokaryotic type I DNA topoisomerase70 d2j0sc1
not modelled
19.1
63
Fold: Mago nashi proteinSuperfamily: Mago nashi proteinFamily: Mago nashi protein71 c1nyjC_
not modelled
18.7
26
PDB header: viral proteinChain: C: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
72 c1nyjD_
not modelled
18.7
26
PDB header: viral proteinChain: D: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
73 c2kqtC_
not modelled
18.7
26
PDB header: transport proteinChain: C: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
74 c2kqtA_
not modelled
18.7
26
PDB header: transport proteinChain: A: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
75 c2kqtB_
not modelled
18.7
26
PDB header: transport proteinChain: B: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
76 c1nyjB_
not modelled
18.7
26
PDB header: viral proteinChain: B: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
77 c1mp6A_
not modelled
18.7
26
PDB header: membrane proteinChain: A: PDB Molecule: matrix protein m2;PDBTitle: structure of the transmembrane region of the m2 protein h+2 channel by solid state nmr spectroscopy
78 c2kqtD_
not modelled
18.7
26
PDB header: transport proteinChain: D: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
79 c1nyjA_
not modelled
18.7
26
PDB header: viral proteinChain: A: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
80 c3ooxA_
not modelled
18.0
21
PDB header: oxidoreductaseChain: A: PDB Molecule: putative 2og-fe(ii) oxygenase family protein;PDBTitle: crystal structure of a putative 2og-fe(ii) oxygenase family protein2 (cc_0200) from caulobacter crescentus at 1.44 a resolution
81 c2ph7B_
not modelled
18.0
19
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein af_2093;PDBTitle: crystal structure of af2093 from archaeoglobus fulgidus
82 c3memA_
not modelled
17.5
21
PDB header: signaling proteinChain: A: PDB Molecule: putative signal transduction protein;PDBTitle: crystal structure of a putative signal transduction protein2 (maqu_0641) from marinobacter aquaeolei vt8 at 2.25 a resolution
83 c2gajA_
not modelled
17.2
13
PDB header: isomeraseChain: A: PDB Molecule: dna topoisomerase i;PDBTitle: structure of full length topoisomerase i from thermotoga maritima in2 monoclinic crystal form
84 c2kadA_
not modelled
15.3
26
PDB header: membrane proteinChain: A: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
85 c2kadD_
not modelled
15.3
26
PDB header: membrane proteinChain: D: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
86 c2kadC_
not modelled
15.3
26
PDB header: membrane proteinChain: C: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
87 c2kadB_
not modelled
15.3
26
PDB header: membrane proteinChain: B: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
88 d2gycw1
not modelled
14.2
14
Fold: Long alpha-hairpinSuperfamily: Ribosomal protein L29 (L29p)Family: Ribosomal protein L29 (L29p)89 c2jmxA_
not modelled
14.0
15
PDB header: hydrolaseChain: A: PDB Molecule: atp synthase o subunit, mitochondrial;PDBTitle: oscp-nt (1-120) in complex with n-terminal (1-25) alpha2 subunit from f1-atpase
90 d1x9na2
not modelled
13.9
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA ligase/mRNA capping enzyme postcatalytic domain91 c2zkrv_
not modelled
13.7
20
PDB header: ribosomal protein/rnaChain: V: PDB Molecule: rna expansion segment es9 part2;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
92 d1gw5b_
not modelled
13.5
16
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: Clathrin adaptor core protein93 c3kaeC_
not modelled
13.3
28
PDB header: protein bindingChain: C: PDB Molecule: possible protein of nuclear scaffold;PDBTitle: cdc27 n-terminus
94 c3c9jD_
not modelled
12.8
21
PDB header: membrane proteinChain: D: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
95 c3c9jA_
not modelled
12.8
21
PDB header: membrane proteinChain: A: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
96 c3c9jB_
not modelled
12.8
21
PDB header: membrane proteinChain: B: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
97 c3c9jC_
not modelled
12.8
21
PDB header: membrane proteinChain: C: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
98 c2g9tT_
not modelled
12.6
54
PDB header: viral proteinChain: T: PDB Molecule: PDBTitle: crystal structure of the sars coronavirus nsp10 at 2.1a
99 d1wgla_
not modelled
12.4
11
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: CUE domain