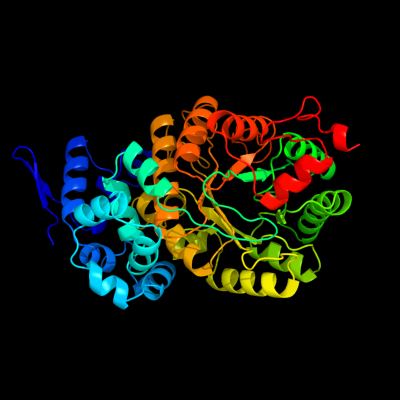
1 c3abqA_
100.0
100
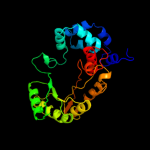
PDB header: lyaseChain: A: PDB Molecule: ethanolamine ammonia-lyase heavy chain;PDBTitle: crystal structure of ethanolamine ammonia-lyase from escherichia coli2 complexed with cn-cbl and 2-amino-1-propanol
2 c2qezC_
100.0
60
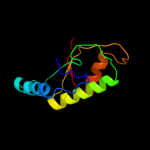
PDB header: lyaseChain: C: PDB Molecule: ethanolamine ammonia-lyase heavy chain;PDBTitle: crystal structure of ethanolamine ammonia-lyase heavy chain2 (yp_013784.1) from listeria monocytogenes 4b f2365 at 2.15 a3 resolution
3 d1eexa_
94.6
16
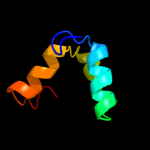
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: Diol dehydratase, alpha subunit4 d1iwpa_
93.9
18
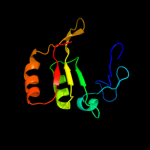
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: Diol dehydratase, alpha subunit5 c3bwwA_
53.4
16
PDB header: metal binding proteinChain: A: PDB Molecule: protein of unknown function duf692/cog3220;PDBTitle: crystal structure of a duf692 family protein (hs_1138) from2 haemophilus somnus 129pt at 2.20 a resolution
6 c3bq7A_
51.1
16
PDB header: transferaseChain: A: PDB Molecule: diacylglycerol kinase delta;PDBTitle: sam domain of diacylglycerol kinase delta1 (e35g)
7 c2fpgA_
43.0
22
PDB header: ligaseChain: A: PDB Molecule: succinyl-coa ligase [gdp-forming] alpha-chain,PDBTitle: crystal structure of pig gtp-specific succinyl-coa2 synthetase in complex with gdp
8 c1v85A_
39.8
10
PDB header: apoptosisChain: A: PDB Molecule: similar to ring finger protein 36;PDBTitle: sterile alpha motif (sam) domain of mouse bifunctional2 apoptosis regulator
9 c2nu8D_
37.2
21
PDB header: ligaseChain: D: PDB Molecule: succinyl-coa ligase [adp-forming] subunit alpha;PDBTitle: c123at mutant of e. coli succinyl-coa synthetase
10 c3qmlC_
35.9
17
PDB header: chaperone/protein transportChain: C: PDB Molecule: nucleotide exchange factor sil1;PDBTitle: the structural analysis of sil1-bip complex reveals the mechanism for2 sil1 to function as a novel nucleotide exchange factor
11 d2csua1
35.9
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain12 c3fokH_
32.5
11
PDB header: structural genomics, unknown functionChain: H: PDB Molecule: uncharacterized protein cgl0159;PDBTitle: crystal structure of cgl0159 from corynebacterium2 glutamicum (brevibacterium flavum). northeast structural3 genomics target cgr115
13 c3njbA_
30.5
10
PDB header: lyaseChain: A: PDB Molecule: enoyl-coa hydratase;PDBTitle: crystal structure of enoyl-coa hydratase from mycobacterium smegmatis,2 iodide soak
14 c3oqhB_
28.6
19
PDB header: ligaseChain: B: PDB Molecule: putative uncharacterized protein yvmc;PDBTitle: crystal structure of b. licheniformis cdps yvmc-blic
15 c2bh7A_
28.1
15
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylmuramoyl-l-alanine amidase;PDBTitle: crystal structure of a semet derivative of amid at 2.22 angstroms
16 c2duwA_
28.0
28
PDB header: ligand binding proteinChain: A: PDB Molecule: putative coa-binding protein;PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
17 d1j3ba1
26.5
18
Fold: PEP carboxykinase-likeSuperfamily: PEP carboxykinase-likeFamily: PEP carboxykinase C-terminal domain18 c3pcsB_
26.2
27
PDB header: protein transport/transferaseChain: B: PDB Molecule: espg;PDBTitle: structure of espg-pak2 autoinhibitory ialpha3 helix complex
19 c3qd5B_
23.7
24
PDB header: isomeraseChain: B: PDB Molecule: putative ribose-5-phosphate isomerase;PDBTitle: crystal structure of a putative ribose-5-phosphate isomerase from2 coccidioides immitis solved by combined iodide ion sad and mr
20 d1iuka_
21.7
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain21 d1o1xa_
not modelled
21.1
22
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB22 c1ye9E_
not modelled
20.5
25
PDB header: oxidoreductaseChain: E: PDB Molecule: catalase hpii;PDBTitle: crystal structure of proteolytically truncated catalase2 hpii from e. coli
23 c3bs5A_
not modelled
20.4
14
PDB header: signaling protein/membrane proteinChain: A: PDB Molecule: protein aveugle;PDBTitle: crystal structure of hcnk2-sam/dhyp-sam complex
24 d1pk1c1
not modelled
18.8
21
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain25 c2iufA_
not modelled
18.2
25
PDB header: oxidoreductaseChain: A: PDB Molecule: catalase;PDBTitle: the structures of penicillium vitale catalase: resting2 state, oxidised state (compound i) and complex with3 aminotriazole
26 d1a4ea_
not modelled
18.2
17
Fold: Heme-dependent catalase-likeSuperfamily: Heme-dependent catalase-likeFamily: Heme-dependent catalases27 c1zcjA_
not modelled
17.8
23
PDB header: oxidoreductaseChain: A: PDB Molecule: peroxisomal bifunctional enzyme;PDBTitle: crystal structure of 3-hydroxyacyl-coa dehydrogenase
28 d1y81a1
not modelled
17.6
37
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain29 c3oqvA_
not modelled
17.4
27
PDB header: protein bindingChain: A: PDB Molecule: albc;PDBTitle: albc, a cyclodipeptide synthase from streptomyces noursei
30 d1si8a_
not modelled
17.0
25
Fold: Heme-dependent catalase-likeSuperfamily: Heme-dependent catalase-likeFamily: Heme-dependent catalases31 d1sqia2
not modelled
17.0
19
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases32 d1e93a_
not modelled
16.9
25
Fold: Heme-dependent catalase-likeSuperfamily: Heme-dependent catalase-likeFamily: Heme-dependent catalases33 d1dkua1
not modelled
16.5
18
Fold: PRTase-likeSuperfamily: PRTase-likeFamily: Phosphoribosylpyrophosphate synthetase-like34 d1gwea_
not modelled
16.5
38
Fold: Heme-dependent catalase-likeSuperfamily: Heme-dependent catalase-likeFamily: Heme-dependent catalases35 d4blca_
not modelled
16.4
33
Fold: Heme-dependent catalase-likeSuperfamily: Heme-dependent catalase-likeFamily: Heme-dependent catalases36 d1p80a2
not modelled
16.4
25
Fold: Heme-dependent catalase-likeSuperfamily: Heme-dependent catalase-likeFamily: Heme-dependent catalases37 c3ej6D_
not modelled
16.4
25
PDB header: oxidoreductaseChain: D: PDB Molecule: catalase-3;PDBTitle: neurospora crassa catalase-3 crystal structure
38 d1qwla_
not modelled
16.2
29
Fold: Heme-dependent catalase-likeSuperfamily: Heme-dependent catalase-likeFamily: Heme-dependent catalases39 d1otha2
not modelled
16.1
10
Fold: ATC-likeSuperfamily: Aspartate/ornithine carbamoyltransferaseFamily: Aspartate/ornithine carbamoyltransferase40 c3lq1A_
not modelled
16.0
21
PDB header: transferaseChain: A: PDB Molecule: 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-PDBTitle: crystal structure of 2-succinyl-6-hydroxy-2,4-cyclohexadiene2 1-carboxylic acid synthase/2-oxoglutarate decarboxylase3 from listeria monocytogenes str. 4b f2365
41 c2pc9B_
not modelled
15.6
21
PDB header: lyaseChain: B: PDB Molecule: phosphoenolpyruvate carboxykinase [atp];PDBTitle: crystal structure of atp-dependent phosphoenolpyruvate carboxykinase2 from thermus thermophilus hb8
42 c1sy7B_
not modelled
15.5
29
PDB header: oxidoreductaseChain: B: PDB Molecule: catalase 1;PDBTitle: crystal structure of the catalase-1 from neurospora crassa, native2 structure at 1.75a resolution.
43 d1mj3a_
not modelled
15.2
20
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Crotonase-like44 d1m7sa_
not modelled
15.1
25
Fold: Heme-dependent catalase-likeSuperfamily: Heme-dependent catalase-likeFamily: Heme-dependent catalases45 c2j2mD_
not modelled
14.7
29
PDB header: oxidoreductaseChain: D: PDB Molecule: catalase;PDBTitle: crystal structure analysis of catalase from exiguobacterium2 oxidotolerans
46 c2kp7A_
not modelled
14.4
50
PDB header: hydrolaseChain: A: PDB Molecule: crossover junction endonuclease mus81;PDBTitle: solution nmr structure of the mus81 n-terminal hhh.2 northeast structural genomics consortium target mmt1a
47 d1ofua2
not modelled
14.2
32
Fold: Bacillus chorismate mutase-likeSuperfamily: Tubulin C-terminal domain-likeFamily: Tubulin, C-terminal domain48 c2dhmA_
not modelled
14.2
10
PDB header: protein bindingChain: A: PDB Molecule: protein bola;PDBTitle: solution structure of the bola protein from escherichia coli
49 c1p81A_
not modelled
13.7
25
PDB header: oxidoreductaseChain: A: PDB Molecule: catalase hpii;PDBTitle: crystal structure of the d181e variant of catalase hpii2 from e. coli
50 c3tr3A_
not modelled
13.6
15
PDB header: unknown functionChain: A: PDB Molecule: bola;PDBTitle: structure of a bola protein homologue from coxiella burnetii
51 c2xq1M_
not modelled
13.6
29
PDB header: oxidoreductaseChain: M: PDB Molecule: peroxisomal catalase;PDBTitle: crystal structure of peroxisomal catalase from the yeast hansenula2 polymorpha
52 d1dgfa_
not modelled
13.6
33
Fold: Heme-dependent catalase-likeSuperfamily: Heme-dependent catalase-likeFamily: Heme-dependent catalases53 c1xs3A_
not modelled
13.6
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein xc975;PDBTitle: solution structure analysis of the xc975 protein
54 c3mogA_
not modelled
13.4
19
PDB header: oxidoreductaseChain: A: PDB Molecule: probable 3-hydroxybutyryl-coa dehydrogenase;PDBTitle: crystal structure of 3-hydroxybutyryl-coa dehydrogenase from2 escherichia coli k12 substr. mg1655
55 d2ae9a1
not modelled
13.1
32
Fold: DNA polymerase III theta subunit-likeSuperfamily: DNA polymerase III theta subunit-likeFamily: DNA polymerase III theta subunit-like56 c2gleA_
not modelled
12.9
11
PDB header: protein bindingChain: A: PDB Molecule: neurabin-1;PDBTitle: solution structure of neurabin sam domain
57 d1kw4a_
not modelled
12.7
13
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain58 c1f4jC_
not modelled
12.5
33
PDB header: oxidoreductaseChain: C: PDB Molecule: catalase;PDBTitle: structure of tetragonal crystals of human erythrocyte2 catalase
59 d1nxha_
not modelled
12.1
15
Fold: Hypothetical protein MTH393Superfamily: Hypothetical protein MTH393Family: Hypothetical protein MTH39360 c3imoC_
not modelled
12.1
42
PDB header: unknown functionChain: C: PDB Molecule: integron cassette protein;PDBTitle: structure from the mobile metagenome of vibrio cholerae.2 integron cassette protein vch_cass14
61 d2f6qa1
not modelled
11.9
17
Fold: ClpP/crotonaseSuperfamily: ClpP/crotonaseFamily: Crotonase-like62 c1pk1A_
not modelled
11.7
11
PDB header: transcription repressionChain: A: PDB Molecule: polyhomeotic-proximal chromatin protein;PDBTitle: hetero sam domain structure of ph and scm.
63 c3onoA_
not modelled
11.5
19
PDB header: isomeraseChain: A: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of ribose-5-phosphate isomerase lacab_rpib from2 vibrio parahaemolyticus
64 d1yzya1
not modelled
11.5
16
Fold: YgbK-likeSuperfamily: YgbK-likeFamily: YgbK-like65 d2f2aa1
not modelled
11.0
20
Fold: Amidase signature (AS) enzymesSuperfamily: Amidase signature (AS) enzymesFamily: Amidase signature (AS) enzymes66 c2pzlB_
not modelled
10.9
15
PDB header: sugar binding proteinChain: B: PDB Molecule: putative nucleotide sugar epimerase/ dehydratase;PDBTitle: crystal structure of the bordetella bronchiseptica enzyme2 wbmg in complex with nad and udp
67 d1ny8a_
not modelled
10.6
15
Fold: Alpha-lytic protease prodomain-likeSuperfamily: BolA-likeFamily: BolA-like68 c3fksY_
not modelled
10.6
16
PDB header: hydrolaseChain: Y: PDB Molecule: atp synthase subunit gamma, mitochondrial;PDBTitle: yeast f1 atpase in the absence of bound nucleotides
69 d1wwva1
not modelled
10.1
7
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain70 c3c5yD_
not modelled
10.0
30
PDB header: isomeraseChain: D: PDB Molecule: ribose/galactose isomerase;PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
71 d2idob1
not modelled
9.5
29
Fold: DNA polymerase III theta subunit-likeSuperfamily: DNA polymerase III theta subunit-likeFamily: DNA polymerase III theta subunit-like72 c3bs7A_
not modelled
9.5
14
PDB header: signaling proteinChain: A: PDB Molecule: protein aveugle;PDBTitle: crystal structure of the sterile alpha motif (sam) domain2 of hyphen/aveugle
73 c2la7A_
not modelled
9.5
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of the protein yp_557733.1 from burkholderia xenovorans
74 c2ppwA_
not modelled
9.3
17
PDB header: isomeraseChain: A: PDB Molecule: conserved domain protein;PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
75 d1chka_
not modelled
9.2
17
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: Chitosanase76 c3dqqB_
not modelled
9.0
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative trna synthase;PDBTitle: the crystal structure of the putative trna synthase from salmonella2 typhimurium lt2
77 d1ucua_
not modelled
8.9
16
Fold: Phase 1 flagellinSuperfamily: Phase 1 flagellinFamily: Phase 1 flagellin78 d2bgxa2
not modelled
8.9
15
Fold: N-acetylmuramoyl-L-alanine amidase-likeSuperfamily: N-acetylmuramoyl-L-alanine amidase-likeFamily: N-acetylmuramoyl-L-alanine amidase-like79 c2dc0A_
not modelled
8.7
10
PDB header: hydrolaseChain: A: PDB Molecule: probable amidase;PDBTitle: crystal structure of amidase
80 c2d3tB_
not modelled
8.6
28
PDB header: lyase, oxidoreductase/transferaseChain: B: PDB Molecule: fatty oxidation complex alpha subunit;PDBTitle: fatty acid beta-oxidation multienzyme complex from2 pseudomonas fragi, form v
81 c2h2mA_
not modelled
8.4
16
PDB header: metal transportChain: A: PDB Molecule: comm domain-containing protein 1;PDBTitle: solution structure of the n-terminal domain of commd12 (murr1)
82 c2wtbA_
not modelled
8.3
25
PDB header: oxidoreductaseChain: A: PDB Molecule: fatty acid multifunctional protein (atmfp2);PDBTitle: arabidopsis thaliana multifuctional protein, mfp2
83 c1v60A_
not modelled
8.2
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: riken cdna 1810037g04;PDBTitle: solution structure of bola1 protein from mus musculus
84 c3lfmA_
not modelled
8.1
28
PDB header: oxidoreductaseChain: A: PDB Molecule: protein fto;PDBTitle: crystal structure of the fat mass and obesity associated (fto) protein2 reveals basis for its substrate specificity
85 c3ayhB_
not modelled
8.0
12
PDB header: transcriptionChain: B: PDB Molecule: dna-directed rna polymerase iii subunit rpc8;PDBTitle: crystal structure of the c17/25 subcomplex from s. pombe rna2 polymerase iii
86 d1se7a_
not modelled
7.7
29
Fold: DNA polymerase III theta subunit-likeSuperfamily: DNA polymerase III theta subunit-likeFamily: DNA polymerase III theta subunit-like87 c2wa0A_
not modelled
7.6
33
PDB header: immune systemChain: A: PDB Molecule: melanoma-associated antigen 4;PDBTitle: crystal structure of the human magea4
88 d1t47a2
not modelled
7.5
23
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases89 d1vmja_
not modelled
7.5
24
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like90 d2vvpa1
not modelled
7.5
16
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB91 c2fvuA_
not modelled
7.5
50
PDB header: transcriptionChain: A: PDB Molecule: regulatory protein sir3;PDBTitle: structure of the yeast sir3 bah domain
92 d1bqva_
not modelled
7.4
18
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: Pointed domain93 c3n5lA_
not modelled
7.3
7
PDB header: transport proteinChain: A: PDB Molecule: binding protein component of abc phosphonate transporter;PDBTitle: crystal structure of a binding protein component of abc phosphonate2 transporter (pa3383) from pseudomonas aeruginosa at 1.97 a resolution
94 c3gebC_
not modelled
7.1
27
PDB header: hydrolaseChain: C: PDB Molecule: eyes absent homolog 2;PDBTitle: crystal structure of edeya2
95 c2yv1A_
not modelled
7.1
19
PDB header: ligaseChain: A: PDB Molecule: succinyl-coa ligase [adp-forming] subunit alpha;PDBTitle: crystal structure of succinyl-coa synthetase alpha chain from2 methanocaldococcus jannaschii dsm 2661
96 c2kveA_
not modelled
6.8
19
PDB header: hormoneChain: A: PDB Molecule: mesencephalic astrocyte-derived neurotrophic factor;PDBTitle: c-terminal domain of mesencephalic astrocyte-derived neurotrophic2 factor (manf)
97 d1sp9a_
not modelled
6.8
23
Fold: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseSuperfamily: Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenaseFamily: Extradiol dioxygenases98 d1b33b_
not modelled
6.7
25
Fold: Globin-likeSuperfamily: Globin-likeFamily: Phycocyanin-like phycobilisome proteins99 d1u1ha1
not modelled
6.7
24
Fold: TIM beta/alpha-barrelSuperfamily: UROD/MetE-likeFamily: Cobalamin-independent methionine synthase