| 1 | c4a1oB_ |
|
 |
100.0 |
46 |
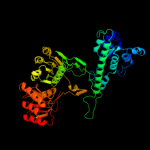
PDB header:transferase-hydrolase
Chain: B: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of mycobacterium tuberculosis purh complexed with2 aicar and a novel nucleotide cfair, at 2.48 a resolution.
|
| 2 | c1thzA_ |
|
 |
100.0 |
42 |
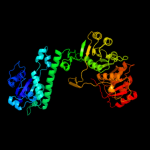
PDB header:transferase, hydrolase
Chain: A: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of avian aicar transformylase in complex2 with a novel inhibitor identified by virtual ligand3 screening
|
| 3 | c1zczA_ |
|
 |
100.0 |
40 |
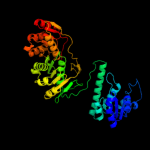
PDB header:transferase/hydrolase
Chain: A: PDB Molecule:bifunctional purine biosynthesis protein purh;
PDBTitle: crystal structure of phosphoribosylaminoimidazolecarboxamide2 formyltransferase / imp cyclohydrolase (tm1249) from thermotoga3 maritima at 1.88 a resolution
|
| 4 | d1g8ma2 |
|
 |
100.0 |
37 |
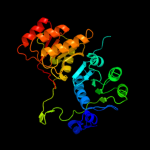
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC |
| 5 | d1pkxa2 |
|
 |
100.0 |
42 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC |
| 6 | d1zcza2 |
|
 |
100.0 |
42 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC |
| 7 | d1pkxa1 |
|
 |
100.0 |
51 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 8 | d1g8ma1 |
|
 |
100.0 |
48 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 9 | d1zcza1 |
|
 |
100.0 |
39 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 10 | d1a9xa2 |
|
 |
99.4 |
25 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain |
| 11 | c2yvqA_ |
|
 |
99.3 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
| 12 | d1wo8a1 |
|
 |
98.3 |
23 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 13 | c1m6vE_ |
|
 |
98.0 |
44 |
PDB header:ligase
Chain: E: PDB Molecule:carbamoyl phosphate synthetase large chain;
PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
|
| 14 | d1vmda_ |
|
 |
96.9 |
26 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 15 | c2pjuD_ |
|
 |
89.7 |
12 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
|
| 16 | c3ff4A_ |
|
 |
89.1 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein chu_1412
|
| 17 | d1iuka_ |
|
 |
88.0 |
35 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 18 | d2d59a1 |
|
 |
85.4 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 19 | d2pjua1 |
|
 |
85.1 |
11 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
| 20 | c2q5cA_ |
|
 |
82.6 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 21 | d2isba1 |
|
not modelled |
82.5 |
22 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:FumA C-terminal domain-like
Family:FumA C-terminal domain-like |
| 22 | d2ioja1 |
|
not modelled |
81.8 |
24 |
Fold:MurF and HprK N-domain-like
Superfamily:HprK N-terminal domain-like
Family:DRTGG domain |
| 23 | d1wbha1 |
|
not modelled |
79.5 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 24 | d2vzsa5 |
|
not modelled |
77.8 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 25 | c2duwA_ |
|
not modelled |
76.1 |
18 |
PDB header:ligand binding protein
Chain: A: PDB Molecule:putative coa-binding protein;
PDBTitle: solution structure of putative coa-binding protein of2 klebsiella pneumoniae
|
| 26 | d1y81a1 |
|
not modelled |
75.8 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 27 | d1b93a_ |
|
not modelled |
75.0 |
21 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Methylglyoxal synthase, MgsA |
| 28 | d1ko7a1 |
|
not modelled |
71.4 |
15 |
Fold:MurF and HprK N-domain-like
Superfamily:HprK N-terminal domain-like
Family:HPr kinase/phoshatase HprK N-terminal domain |
| 29 | c2yw3E_ |
|
not modelled |
70.3 |
34 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-
PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
|
| 30 | c2hvwC_ |
|
not modelled |
69.6 |
23 |
PDB header:hydrolase
Chain: C: PDB Molecule:deoxycytidylate deaminase;
PDBTitle: crystal structure of dcmp deaminase from streptococcus2 mutans
|
| 31 | d1mxsa_ |
|
not modelled |
68.4 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 32 | c2o7pA_ |
|
not modelled |
64.6 |
28 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:riboflavin biosynthesis protein ribd;
PDBTitle: the crystal structure of ribd from escherichia coli in complex with2 the oxidised nadp+ cofactor in the active site of the reductase3 domain
|
| 33 | d1mkza_ |
|
not modelled |
63.8 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 34 | d1vhca_ |
|
not modelled |
63.1 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 35 | d1qt1a_ |
|
not modelled |
62.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 36 | c2e3zB_ |
|
not modelled |
61.7 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of intracellular family 1 beta-2 glucosidase bgl1a from the basidiomycete phanerochaete3 chrysosporium in substrate-free form
|
| 37 | c1ko7B_ |
|
not modelled |
60.7 |
15 |
PDB header:transferase,hydrolase
Chain: B: PDB Molecule:hpr kinase/phosphatase;
PDBTitle: x-ray structure of the hpr kinase/phosphatase from2 staphylococcus xylosus at 1.95 a resolution
|
| 38 | d2obba1 |
|
not modelled |
60.1 |
17 |
Fold:HAD-like
Superfamily:HAD-like
Family:BT0820-like |
| 39 | d1ytla1 |
|
not modelled |
59.5 |
17 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:ACDE2-like |
| 40 | c3ia7A_ |
|
not modelled |
58.4 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:calg4;
PDBTitle: crystal structure of calg4, the calicheamicin glycosyltransferase
|
| 41 | d2csua1 |
|
not modelled |
57.0 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 42 | d1ujpa_ |
|
not modelled |
55.7 |
30 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 43 | d1f75a_ |
|
not modelled |
55.5 |
14 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
| 44 | d1f0ka_ |
|
not modelled |
55.0 |
26 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Peptidoglycan biosynthesis glycosyltransferase MurG |
| 45 | c3u7jA_ |
|
not modelled |
54.7 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
|
| 46 | c3dmyA_ |
|
not modelled |
52.6 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein fdra;
PDBTitle: crystal structure of a predicated acyl-coa synthetase from e.coli
|
| 47 | d1sjpa2 |
|
not modelled |
52.3 |
21 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 48 | c3e2vA_ |
|
not modelled |
51.9 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:3'-5'-exonuclease;
PDBTitle: crystal structure of an uncharacterized amidohydrolase from2 saccharomyces cerevisiae
|
| 49 | d1cbga_ |
|
not modelled |
51.7 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 50 | d1y5ea1 |
|
not modelled |
51.0 |
21 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 51 | d2b3za2 |
|
not modelled |
50.2 |
19 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
| 52 | d1ueha_ |
|
not modelled |
49.8 |
28 |
Fold:Undecaprenyl diphosphate synthase
Superfamily:Undecaprenyl diphosphate synthase
Family:Undecaprenyl diphosphate synthase |
| 53 | d1we3a2 |
|
not modelled |
46.8 |
22 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 54 | c3p9zA_ |
|
not modelled |
44.5 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:uroporphyrinogen iii cosynthase (hemd);
PDBTitle: crystal structure of uroporphyrinogen-iii synthetase from helicobacter2 pylori 26695
|
| 55 | c1xtzA_ |
|
not modelled |
44.2 |
25 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: crystal structure of the s. cerevisiae d-ribose-5-phosphate isomerase:2 comparison with the archeal and bacterial enzymes
|
| 56 | d1pn3a_ |
|
not modelled |
43.3 |
19 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Gtf glycosyltransferase |
| 57 | c3orgB_ |
|
not modelled |
42.8 |
11 |
PDB header:transport protein
Chain: B: PDB Molecule:cmclc;
PDBTitle: crystal structure of a eukaryotic clc transporter
|
| 58 | c3ahxC_ |
|
not modelled |
42.7 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-glucosidase a;
PDBTitle: crystal structure of beta-glucosidase a from bacterium clostridium2 cellulovorans
|
| 59 | d1rd5a_ |
|
not modelled |
42.1 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
| 60 | c2v82A_ |
|
not modelled |
42.0 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
| 61 | c3labA_ |
|
not modelled |
41.9 |
30 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative kdpg (2-keto-3-deoxy-6-phosphogluconate)
PDBTitle: crystal structure of a putative kdpg (2-keto-3-deoxy-6-2 phosphogluconate) aldolase from oleispira antarctica
|
| 62 | d1tz9a_ |
|
not modelled |
41.4 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:UxuA-like |
| 63 | c1nh7A_ |
|
not modelled |
40.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:atp phosphoribosyltransferase;
PDBTitle: atp phosphoribosyltransferase (atp-prtase) from mycobacterium2 tuberculosis
|
| 64 | c3sajB_ |
|
not modelled |
40.7 |
10 |
PDB header:transport protein
Chain: B: PDB Molecule:glutamate receptor 1;
PDBTitle: crystal structure of glutamate receptor glua1 amino terminal domain
|
| 65 | d2pq6a1 |
|
not modelled |
40.4 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
| 66 | d1zpva1 |
|
not modelled |
40.2 |
27 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:SP0238-like |
| 67 | c2z1sA_ |
|
not modelled |
40.2 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase b;
PDBTitle: beta-glucosidase b from paenibacillus polymyxa complexed2 with cellotetraose
|
| 68 | d1bqca_ |
|
not modelled |
39.5 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 69 | d1rrva_ |
|
not modelled |
39.4 |
23 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Gtf glycosyltransferase |
| 70 | d1kida_ |
|
not modelled |
39.3 |
24 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 71 | c3ktcB_ |
|
not modelled |
39.3 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:xylose isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
|
| 72 | c3fuyC_ |
|
not modelled |
39.2 |
35 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:putative integron gene cassette protein;
PDBTitle: structure from the mobile metagenome of cole harbour salt2 marsh: integron cassette protein hfx_cass1
|
| 73 | d1k77a_ |
|
not modelled |
38.3 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Hypothetical protein YgbM (EC1530) |
| 74 | c3ahyD_ |
|
not modelled |
38.1 |
25 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of beta-glucosidase 2 from fungus trichoderma reesei2 in complex with tris
|
| 75 | c2f00A_ |
|
not modelled |
37.3 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate--l-alanine ligase;
PDBTitle: escherichia coli murc
|
| 76 | d1vffa1 |
|
not modelled |
37.2 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 77 | c2hxvA_ |
|
not modelled |
36.5 |
18 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:diaminohydroxyphosphoribosylaminopyrimidine deaminase/ 5-
PDBTitle: crystal structure of a diaminohydroxyphosphoribosylaminopyrimidine2 deaminase/ 5-amino-6-(5-phosphoribosylamino)uracil reductase (tm1828)3 from thermotoga maritima at 1.80 a resolution
|
| 78 | c2csuB_ |
|
not modelled |
35.6 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:457aa long hypothetical protein;
PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
|
| 79 | c2vldA_ |
|
not modelled |
35.4 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:upf0286 protein pyrab01260;
PDBTitle: crystal structure of a repair endonuclease from pyrococcus2 abyssi
|
| 80 | d1xima_ |
|
not modelled |
34.8 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 81 | d1wcga1 |
|
not modelled |
34.6 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
| 82 | d1xsza2 |
|
not modelled |
33.2 |
39 |
Fold:TBP-like
Superfamily:RalF, C-terminal domain
Family:RalF, C-terminal domain |
| 83 | c2b8eB_ |
|
not modelled |
32.6 |
11 |
PDB header:membrane protein
Chain: B: PDB Molecule:cation-transporting atpase;
PDBTitle: copa atp binding domain
|
| 84 | d1ht6a2 |
|
not modelled |
32.4 |
38 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
| 85 | d1euca1 |
|
not modelled |
32.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 86 | c2pjmA_ |
|
not modelled |
32.0 |
29 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: structure of ribose 5-phosphate isomerase a from2 methanocaldococcus jannaschii
|
| 87 | d1yrra2 |
|
not modelled |
31.7 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:N-acetylglucosamine-6-phosphate deacetylase, NagA, catalytic domain |
| 88 | d1uuqa_ |
|
not modelled |
31.3 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
| 89 | c1uz4A_ |
|
not modelled |
31.3 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:man5a;
PDBTitle: common inhibition of beta-glucosidase and beta-mannosidase2 by isofagomine lactam reflects different conformational3 intineraries for glucoside and mannoside hydrolysis
|
| 90 | c3ptkB_ |
|
not modelled |
31.1 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase os4bglu12;
PDBTitle: the crystal structure of rice (oryza sativa l.) os4bglu12
|
| 91 | c3kg2A_ |
|
not modelled |
31.0 |
4 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:glutamate receptor 2;
PDBTitle: ampa subtype ionotropic glutamate receptor in complex with competitive2 antagonist zk 200775
|
| 92 | d2nu7a1 |
|
not modelled |
30.7 |
34 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
| 93 | c2wjxA_ |
|
not modelled |
30.2 |
4 |
PDB header:transport protein
Chain: A: PDB Molecule:glutamate receptor 2;
PDBTitle: crystal structure of the human ionotropic glutamate2 receptor glur2 atd region at 4.1 a resolution
|
| 94 | c2vzvB_ |
|
not modelled |
30.2 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:exo-beta-d-glucosaminidase;
PDBTitle: substrate complex of amycolatopsis orientalis exo-2 chitosanase csxa e541a with chitosan
|
| 95 | d1snna_ |
|
not modelled |
30.1 |
22 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 96 | c3iv4A_ |
|
not modelled |
29.8 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: a putative oxidoreductase with a thioredoxin fold
|
| 97 | d1yx1a1 |
|
not modelled |
29.4 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:KguE-like |
| 98 | d1g57a_ |
|
not modelled |
29.3 |
32 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
| 99 | c2f8mB_ |
|
not modelled |
29.1 |
31 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: ribose 5-phosphate isomerase from plasmodium falciparum
|
| 100 | d1un7a2 |
|
not modelled |
29.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:N-acetylglucosamine-6-phosphate deacetylase, NagA, catalytic domain |
| 101 | c1i8tB_ |
|
not modelled |
29.0 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-galactopyranose mutase;
PDBTitle: strcuture of udp-galactopyranose mutase from e.coli
|
| 102 | c1gqqA_ |
|
not modelled |
28.9 |
21 |
PDB header:cell wall biosynthesis
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus2 influenzae
|
| 103 | c3u57A_ |
|
not modelled |
28.9 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:raucaffricine-o-beta-d-glucosidase;
PDBTitle: structures of alkaloid biosynthetic glucosidases decode substrate2 specificity
|
| 104 | c2rbgB_ |
|
not modelled |
28.8 |
24 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative uncharacterized protein st0493;
PDBTitle: crystal structure of hypothetical protein(st0493) from2 sulfolobus tokodaii
|
| 105 | d1tg7a5 |
|
not modelled |
28.6 |
30 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Glycosyl hydrolases family 35 catalytic domain |
| 106 | c3qomA_ |
|
not modelled |
28.1 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phospho-beta-glucosidase;
PDBTitle: crystal structure of 6-phospho-beta-glucosidase from lactobacillus2 plantarum
|
| 107 | d1oela2 |
|
not modelled |
27.5 |
24 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 108 | c2je8B_ |
|
not modelled |
27.2 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-mannosidase;
PDBTitle: structure of a beta-mannosidase from bacteroides2 thetaiotaomicron
|
| 109 | d1srva_ |
|
not modelled |
27.0 |
24 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 110 | c2d2rA_ |
|
not modelled |
26.8 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:undecaprenyl pyrophosphate synthase;
PDBTitle: crystal structure of helicobacter pylori undecaprenyl pyrophosphate2 synthase
|
| 111 | c3c2qA_ |
|
not modelled |
26.8 |
35 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized conserved protein;
PDBTitle: crystal structure of conserved putative lor/sdh protein2 from methanococcus maripaludis s2
|
| 112 | c2qyxB_ |
|
not modelled |
26.4 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein mj0159;
PDBTitle: crystal structure of uncharacterized protein mj0159 from2 methanocaldococcus jannaschii
|
| 113 | d2hxva2 |
|
not modelled |
26.1 |
22 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
| 114 | c3mioA_ |
|
not modelled |
25.6 |
41 |
PDB header:lyase
Chain: A: PDB Molecule:3,4-dihydroxy-2-butanone 4-phosphate synthase;
PDBTitle: crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase2 domain from mycobacterium tuberculosis at ph 6.00
|
| 115 | c3kwmC_ |
|
not modelled |
25.4 |
31 |
PDB header:isomerase
Chain: C: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-isomerase a
|
| 116 | d1dk7a_ |
|
not modelled |
25.2 |
28 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:GroEL-like chaperone, apical domain |
| 117 | d1g2qa_ |
|
not modelled |
25.2 |
21 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 118 | d1y5ha3 |
|
not modelled |
24.9 |
12 |
Fold:CBS-domain pair
Superfamily:CBS-domain pair
Family:CBS-domain pair |
| 119 | d2ooda2 |
|
not modelled |
24.8 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:SAH/MTA deaminase-like |
| 120 | d1ceoa_ |
|
not modelled |
24.4 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |

