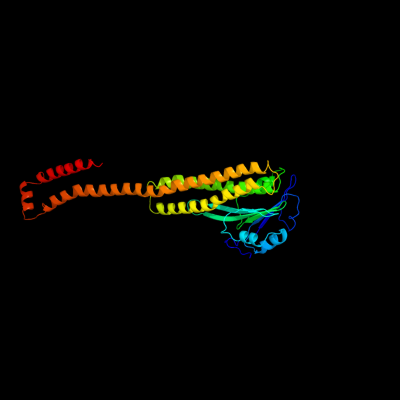
1 c2bbjB_
100.0
15
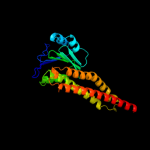
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
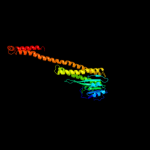
2 c3nwiC_
100.0
90
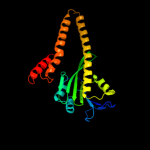
PDB header: transport proteinChain: C: PDB Molecule: zinc transport protein zntb;PDBTitle: the soluble domain structure of the zntb zn2+ efflux system
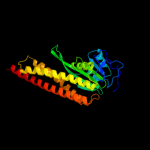
3 d2iuba1
100.0
15
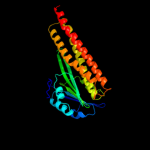
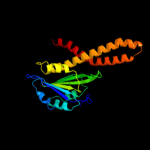
Fold: CorA soluble domain-likeSuperfamily: CorA soluble domain-likeFamily: CorA soluble domain-like4 c3ck6E_
100.0
18
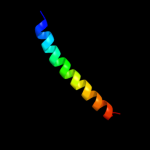
PDB header: structural proteinChain: E: PDB Molecule: putative membrane transport protein;PDBTitle: crystal structure of zntb cytoplasmic domain from vibrio2 parahaemolyticus rimd 2210633
5 c2hn1A_
100.0
16
PDB header: metal transportChain: A: PDB Molecule: magnesium and cobalt transporter;PDBTitle: crystal structure of a cora soluble domain from a. fulgidus in complex2 with co2+
6 d2bbha1
100.0
17
Fold: CorA soluble domain-likeSuperfamily: CorA soluble domain-likeFamily: CorA soluble domain-like7 d2iuba2
99.4
18
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region8 d3cx5d2
64.2
13
Fold: Single transmembrane helixSuperfamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchorFamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor9 d1ppjd2
63.2
19
Fold: Single transmembrane helixSuperfamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchorFamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor10 c3fseB_
33.0
9
PDB header: hydrolaseChain: B: PDB Molecule: two-domain protein containing dj-1/thij/pfpi-like andPDBTitle: crystal structure of a two-domain protein containing dj-1/thij/pfpi-2 like and ferritin-like domains (ava_4496) from anabaena variabilis3 atcc 29413 at 1.90 a resolution
11 c1qcrD_
29.7
19
PDB header: PDB COMPND: 12 c3cwbQ_
27.8
19
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
13 c1p84D_
27.3
13
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
14 c2yiuE_
27.2
6
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c1, heme protein;PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
15 c3dl8H_
26.6
13
PDB header: protein transportChain: H: PDB Molecule: preprotein translocase subunit secy;PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
16 c1eboE_
25.1
16
PDB header: viral proteinChain: E: PDB Molecule: ebola virus envelope protein chimera consistingPDBTitle: crystal structure of the ebola virus membrane-fusion2 subunit, gp2, from the envelope glycoprotein ectodomain
17 c3j01A_
24.7
18
PDB header: ribosome/ribosomal proteinChain: A: PDB Molecule: preprotein translocase secy subunit;PDBTitle: structure of the ribosome-secye complex in the membrane environment
18 c2fynH_
23.7
6
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c1;PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
19 c3ghgK_
21.7
10
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
20 c3tu8A_
21.7
27
PDB header: unknown functionChain: A: PDB Molecule: burkholderia lethal factor 1 (blf1);PDBTitle: crystal structure of the burkholderia lethal factor 1 (blf1)
21 c1zrtD_
not modelled
20.7
9
PDB header: oxidoreductase/metal transportChain: D: PDB Molecule: cytochrome c1;PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
22 c3mkuA_
not modelled
18.7
13
PDB header: transport proteinChain: A: PDB Molecule: multi antimicrobial extrusion protein (na(+)/drugPDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
23 c1s1iJ_
not modelled
18.6
43
PDB header: ribosomeChain: J: PDB Molecule: 60s ribosomal protein l11;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
24 c2akiY_
not modelled
17.9
18
PDB header: protein transportChain: Y: PDB Molecule: preprotein translocase secy subunit;PDBTitle: normal mode-based flexible fitted coordinates of a2 translocating secyeg protein-conducting channel into the3 cryo-em map of a secyeg-nascent chain-70s ribosome complex4 from e. coli
25 c4a1cD_
not modelled
16.0
44
PDB header: ribosomeChain: D: PDB Molecule: 60s ribosomal protein l11;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
26 d1iwga8
not modelled
15.2
11
Fold: Multidrug efflux transporter AcrB transmembrane domainSuperfamily: Multidrug efflux transporter AcrB transmembrane domainFamily: Multidrug efflux transporter AcrB transmembrane domain27 c3rfuC_
not modelled
15.2
10
PDB header: hydrolase, membrane proteinChain: C: PDB Molecule: copper efflux atpase;PDBTitle: crystal structure of a copper-transporting pib-type atpase
28 d1e52a_
not modelled
14.8
27
Fold: Long alpha-hairpinSuperfamily: C-terminal UvrC-binding domain of UvrBFamily: C-terminal UvrC-binding domain of UvrB29 d2d69a2
not modelled
14.7
20
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase30 c1dkgB_
not modelled
14.4
18
PDB header: complex (hsp24/hsp70)Chain: B: PDB Molecule: nucleotide exchange factor grpe;PDBTitle: crystal structure of the nucleotide exchange factor grpe2 bound to the atpase domain of the molecular chaperone dnak
31 c1oy8A_
not modelled
13.6
24
PDB header: membrane proteinChain: A: PDB Molecule: acriflavine resistance protein b;PDBTitle: structural basis of multiple drug binding capacity of the acrb2 multidrug efflux pump
32 d1svda2
not modelled
12.8
33
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase33 c1ei3E_
not modelled
12.8
11
PDB header: PDB COMPND: 34 c2ks1B_
not modelled
12.7
29
PDB header: transferaseChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
35 c2cpbA_
not modelled
12.4
9
PDB header: viral proteinChain: A: PDB Molecule: m13 major coat protein;PDBTitle: solution nmr structures of the major coat protein of2 filamentous bacteriophage m13 solubilized in3 dodecylphosphocholine micelles, 25 lowest energy structures
36 c2akfC_
not modelled
11.6
12
PDB header: protein bindingChain: C: PDB Molecule: coronin-1a;PDBTitle: crystal structure of the coiled-coil domain of coronin 1
37 c2akfB_
not modelled
11.6
12
PDB header: protein bindingChain: B: PDB Molecule: coronin-1a;PDBTitle: crystal structure of the coiled-coil domain of coronin 1
38 c2akfA_
not modelled
11.6
12
PDB header: protein bindingChain: A: PDB Molecule: coronin-1a;PDBTitle: crystal structure of the coiled-coil domain of coronin 1
39 d1bxna2
not modelled
11.5
13
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase40 d1tqga_
not modelled
11.5
14
Fold: Four-helical up-and-down bundleSuperfamily: Histidine-containing phosphotransfer domain, HPT domainFamily: Chemotaxis protein CheA P1 domain41 d1i6za_
not modelled
10.7
6
Fold: Spectrin repeat-likeSuperfamily: BAG domainFamily: BAG domain42 d1wh5a_
not modelled
9.9
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain43 d2p61a1
not modelled
9.6
12
Fold: Four-helical up-and-down bundleSuperfamily: TM1646-likeFamily: TM1646-like44 c2xzm7_
not modelled
9.4
20
PDB header: ribosomeChain: 7: PDB Molecule: plectin/s10 domain containing protein;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
45 c3qthA_
not modelled
9.3
14
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a hypothetical dinb-like protein (cps_3021) from2 colwellia psychrerythraea 34h at 2.20 a resolution
46 c2p61A_
not modelled
9.0
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein tm_1646;PDBTitle: crystal structure of protein tm1646 from thermotoga2 maritima, pfam duf327
47 c3dinF_
not modelled
8.8
18
PDB header: membrane protein, protein transportChain: F: PDB Molecule: preprotein translocase subunit secy;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
48 c2zqpY_
not modelled
8.8
16
PDB header: protein transportChain: Y: PDB Molecule: preprotein translocase secy subunit;PDBTitle: crystal structure of secye translocon from thermus2 thermophilus
49 c1w5kB_
not modelled
8.6
24
PDB header: four helix bundleChain: B: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
50 c1w5kC_
not modelled
8.6
24
PDB header: four helix bundleChain: C: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
51 c1w5kA_
not modelled
8.6
24
PDB header: four helix bundleChain: A: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
52 c1w5kD_
not modelled
8.6
24
PDB header: four helix bundleChain: D: PDB Molecule: general control protein gcn4;PDBTitle: an anti-parallel four helix bundle
53 c3kyiA_
not modelled
8.6
14
PDB header: transferaseChain: A: PDB Molecule: putative histidine protein kinase;PDBTitle: crystal structure of the phosphorylated p1 domain of chea3 in complex2 with chey6 from r. sphaeroides
54 d1rzhh2
not modelled
8.5
36
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region55 d2c2aa1
not modelled
8.5
5
Fold: ROP-likeSuperfamily: Homodimeric domain of signal transducing histidine kinaseFamily: Homodimeric domain of signal transducing histidine kinase56 c2qupA_
not modelled
8.5
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: bh1478 protein;PDBTitle: crystal structure of uncharacterized protein bh1478 from bacillus2 halodurans
57 c3u5gK_
not modelled
8.1
16
PDB header: ribosomeChain: K: PDB Molecule: 40s ribosomal protein s10-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
58 d1l9bh2
not modelled
7.9
36
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region59 c2oszA_
not modelled
7.9
13
PDB header: structural proteinChain: A: PDB Molecule: nucleoporin p58/p45;PDBTitle: structure of nup58/45 suggests flexible nuclear pore diameter by2 intermolecular sliding
60 d1oe4a_
not modelled
7.9
43
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Single-strand selective monofunctional uracil-DNA glycosylase SMUG161 d1qoja_
not modelled
7.7
28
Fold: Long alpha-hairpinSuperfamily: C-terminal UvrC-binding domain of UvrBFamily: C-terminal UvrC-binding domain of UvrB62 c1htjF_
not modelled
7.6
12
PDB header: signaling proteinChain: F: PDB Molecule: kiaa0380;PDBTitle: structure of the rgs-like domain from pdz-rhogef
63 d1htjf_
not modelled
7.6
12
Fold: Regulator of G-protein signaling, RGSSuperfamily: Regulator of G-protein signaling, RGSFamily: Regulator of G-protein signaling, RGS64 d1wpga4
not modelled
7.6
8
Fold: Calcium ATPase, transmembrane domain MSuperfamily: Calcium ATPase, transmembrane domain MFamily: Calcium ATPase, transmembrane domain M65 c2oltB_
not modelled
7.6
13
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a phou-like protein (so_3770) from shewanella2 oneidensis mr-1 at 2.00 a resolution
66 d1wi3a_
not modelled
7.6
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain67 c2y69Y_
not modelled
7.6
13
PDB header: electron transportChain: Y: PDB Molecule: cytochrome c oxidase subunit 7c;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
68 c3c0uA_
not modelled
7.5
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yaeq;PDBTitle: crystal structure of e.coli yaeq protein
69 d1u5pa1
not modelled
7.4
14
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat70 d1mh3a1
not modelled
7.4
4
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain71 c3ixzA_
not modelled
7.3
9
PDB header: hydrolaseChain: A: PDB Molecule: potassium-transporting atpase alpha;PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
72 c2dceA_
not modelled
7.3
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: kiaa1915 protein;PDBTitle: solution structure of the swirm domain of human kiaa19152 protein
73 d1i5na_
not modelled
7.3
14
Fold: Four-helical up-and-down bundleSuperfamily: Histidine-containing phosphotransfer domain, HPT domainFamily: Chemotaxis protein CheA P1 domain74 c2lchA_
not modelled
7.2
15
PDB header: de novo proteinChain: A: PDB Molecule: protein or38;PDBTitle: solution nmr structure of a protein with a redesigned hydrophobic2 core, northeast structural genomics consortium target or38
75 c1s94A_
not modelled
7.2
11
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: s-syntaxin;PDBTitle: crystal structure of the habc domain of neuronal syntaxin from the2 squid loligo pealei
76 d1s94a_
not modelled
7.2
11
Fold: STAT-likeSuperfamily: t-snare proteinsFamily: t-snare proteins77 d1h9aa2
not modelled
7.2
13
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: Glucose 6-phosphate dehydrogenase-like78 c1kmiZ_
not modelled
7.2
9
PDB header: signaling proteinChain: Z: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of an e.coli chemotaxis protein, chez
79 c3qnqD_
not modelled
7.1
20
PDB header: membrane protein, transport proteinChain: D: PDB Molecule: pts system, cellobiose-specific iic component;PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
80 c2wwbA_
not modelled
7.1
11
PDB header: ribosomeChain: A: PDB Molecule: protein transport protein sec61 subunit alpha isoform 1;PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
81 c3r1fO_
not modelled
7.1
11
PDB header: transcriptionChain: O: PDB Molecule: esx-1 secretion-associated regulator espr;PDBTitle: crystal structure of a key regulator of virulence in mycobacterium2 tuberculosis
82 d1uhsa_
not modelled
7.0
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain83 d9anta_
not modelled
7.0
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain84 d1e3oc1
not modelled
6.9
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain85 d1s35a2
not modelled
6.9
8
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat86 d1d6za4
not modelled
6.8
25
Fold: N domain of copper amine oxidase-likeSuperfamily: Copper amine oxidase, domain NFamily: Copper amine oxidase, domain N87 d8ruca2
not modelled
6.8
13
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase88 c2vi6F_
not modelled
6.7
17
PDB header: transcriptionChain: F: PDB Molecule: homeobox protein nanog;PDBTitle: crystal structure of the nanog homeodomain
89 d2rcrh2
not modelled
6.7
36
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region90 c1nfoA_
not modelled
6.7
4
PDB header: lipid transportChain: A: PDB Molecule: apolipoprotein e2;PDBTitle: apolipoprotein e2 (apoe2, d154a mutation)
91 c3movB_
not modelled
6.7
24
PDB header: structural proteinChain: B: PDB Molecule: lamin-b1;PDBTitle: crystal structure of human lamin-b1 coil 2 segment
92 c3b8eC_
not modelled
6.6
13
PDB header: hydrolase/transport proteinChain: C: PDB Molecule: sodium/potassium-transporting atpase subunitPDBTitle: crystal structure of the sodium-potassium pump
93 d1eysh2
not modelled
6.6
25
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region94 c1gk4A_
not modelled
6.5
19
PDB header: vimentinChain: A: PDB Molecule: vimentin;PDBTitle: human vimentin coil 2b fragment (cys2)
95 d1riqa1
not modelled
6.5
12
Fold: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)Superfamily: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)Family: Putative anticodon-binding domain of alanyl-tRNA synthetase (AlaRS)96 d1le8a_
not modelled
6.4
4
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain97 c3a7pB_
not modelled
6.4
15
PDB header: protein transportChain: B: PDB Molecule: autophagy protein 16;PDBTitle: the crystal structure of saccharomyces cerevisiae atg16
98 c2l2tA_
not modelled
6.4
21
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-4;PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
99 d1du0a_
not modelled
6.4
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain