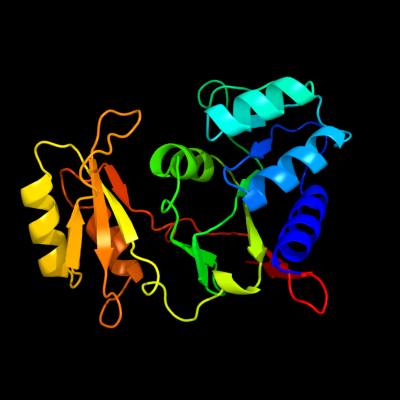
| 1 | c1lkzB_
|
|
 |
100.0 |
97 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase a;
PDBTitle: crystal structure of d-ribose-5-phosphate isomerase (rpia)2 from escherichia coli.
|
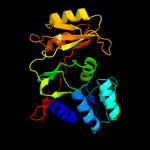
| 2 | c3u7jA_
|
|
 |
100.0 |
67 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
|
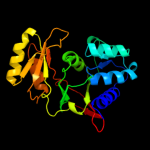
| 3 | c1m0sA_
|
|
 |
100.0 |
64 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: northeast structural genomics consortium (nesg id ir21)
|
| 4 | c3kwmC_
|
|
 |
100.0 |
53 |
PDB header:isomerase
Chain: C: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-isomerase a
|
| 5 | c1lk5C_
|
|
 |
100.0 |
41 |
PDB header:isomerase
Chain: C: PDB Molecule:d-ribose-5-phosphate isomerase;
PDBTitle: structure of the d-ribose-5-phosphate isomerase from2 pyrococcus horikoshii
|
| 6 | c3l7oB_
|
|
 |
100.0 |
41 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
|
| 7 | c1xtzA_
|
|
 |
100.0 |
30 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: crystal structure of the s. cerevisiae d-ribose-5-phosphate isomerase:2 comparison with the archeal and bacterial enzymes
|
| 8 | c3hheA_
|
|
 |
100.0 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from bartonella2 henselae
|
| 9 | c1uj6A_
|
|
 |
100.0 |
41 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of thermus thermophilus ribose-5-phosphate isomerase2 complexed with arabinose-5-phosphate
|
| 10 | c2pjmA_
|
|
 |
100.0 |
43 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: structure of ribose 5-phosphate isomerase a from2 methanocaldococcus jannaschii
|
| 11 | c2f8mB_
|
|
 |
100.0 |
36 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: ribose 5-phosphate isomerase from plasmodium falciparum
|
| 12 | d1o8bb1
|
|
 |
100.0 |
86 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 13 | d1m0sa1
|
|
 |
100.0 |
60 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 14 | d1lk5a1
|
|
 |
100.0 |
34 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 15 | d1uj4a1
|
|
 |
100.0 |
38 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 16 | d1o8ba2
|
|
 |
100.0 |
96 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 17 | d1m0sa2
|
|
 |
99.9 |
68 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 18 | d1lk5a2
|
|
 |
99.9 |
45 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 19 | d1uj4a2
|
|
 |
99.9 |
40 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 20 | d1o8ba1
|
|
 |
99.9 |
63 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:D-ribose-5-phosphate isomerase (RpiA), catalytic domain |
| 21 | c3a11D_ |
|
not modelled |
98.9 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:translation initiation factor eif-2b, delta
PDBTitle: crystal structure of ribose-1,5-bisphosphate isomerase from2 thermococcus kodakaraensis kod1
|
| 22 | c3ecsD_ |
|
not modelled |
98.8 |
15 |
PDB header:translation
Chain: D: PDB Molecule:translation initiation factor eif-2b subunit
PDBTitle: crystal structure of human eif2b alpha
|
| 23 | d1vb5a_ |
|
not modelled |
98.8 |
20 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 24 | d1t5oa_ |
|
not modelled |
98.7 |
16 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 25 | c2yvkA_ |
|
not modelled |
98.7 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of 5-methylthioribose 1-phosphate2 isomerase product complex from bacillus subtilis
|
| 26 | d1t9ka_ |
|
not modelled |
98.7 |
22 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 27 | d2a0ua1 |
|
not modelled |
98.1 |
14 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:IF2B-like |
| 28 | d1poib_ |
|
not modelled |
98.0 |
16 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase beta subunit-like |
| 29 | c3cdkD_ |
|
not modelled |
97.9 |
25 |
PDB header:transferase
Chain: D: PDB Molecule:succinyl-coa:3-ketoacid-coenzyme a transferase
PDBTitle: crystal structure of the co-expressed succinyl-coa2 transferase a and b complex from bacillus subtilis
|
| 30 | c3rrlC_ |
|
not modelled |
97.8 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:succinyl-coa:3-ketoacid-coenzyme a transferase subunit a;
PDBTitle: complex structure of 3-oxoadipate coa-transferase subunit a and b from2 helicobacter pylori 26695
|
| 31 | d2ahua1 |
|
not modelled |
97.8 |
22 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase beta subunit-like |
| 32 | c3cdkA_ |
|
not modelled |
97.7 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:succinyl-coa:3-ketoacid-coenzyme a transferase
PDBTitle: crystal structure of the co-expressed succinyl-coa2 transferase a and b complex from bacillus subtilis
|
| 33 | d1k6da_ |
|
not modelled |
97.7 |
17 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 34 | d1ooya2 |
|
not modelled |
97.6 |
22 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 35 | d1ooya1 |
|
not modelled |
97.5 |
23 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase beta subunit-like |
| 36 | c2ahvC_ |
|
not modelled |
97.4 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:putative enzyme ydif;
PDBTitle: crystal structure of acyl-coa transferase from e. coli o157:h7 (ydif)-2 thioester complex with coa- 1
|
| 37 | d1poia_ |
|
not modelled |
97.4 |
16 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 38 | d1xr4a2 |
|
not modelled |
97.2 |
19 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 39 | c2oasA_ |
|
not modelled |
97.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:4-hydroxybutyrate coenzyme a transferase;
PDBTitle: crystal structure of 4-hydroxybutyrate coenzyme a transferase (atoa)2 in complex with coa from shewanella oneidensis, northeast structural3 genomics target sor119.
|
| 40 | d1xr4a1 |
|
not modelled |
97.0 |
15 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 41 | d2gnpa1 |
|
not modelled |
96.9 |
12 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 42 | c1ooyA_ |
|
not modelled |
96.9 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:succinyl-coa:3-ketoacid-coenzyme a transferase,
PDBTitle: succinyl-coa:3-ketoacid coa transferase from pig heart
|
| 43 | c3gk7A_ |
|
not modelled |
96.8 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:4-hydroxybutyrate coa-transferase;
PDBTitle: crystal structure of 4-hydroxybutyrate coa-transferase from2 clostridium aminobutyricum
|
| 44 | c3eh7A_ |
|
not modelled |
96.8 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:4-hydroxybutyrate coa-transferase;
PDBTitle: the structure of a putative 4-hydroxybutyrate coa-transferase from2 porphyromonas gingivalis w83
|
| 45 | c3kv1A_ |
|
not modelled |
96.8 |
30 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional repressor;
PDBTitle: crystal structure of putative sugar-binding domain of transcriptional2 repressor from vibrio fischeri
|
| 46 | c3nzeB_ |
|
not modelled |
96.6 |
22 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative transcriptional regulator, sugar-binding family;
PDBTitle: the crystal structure of a domain of a possible sugar-binding2 transcriptional regulator from arthrobacter aurescens tc1.
|
| 47 | c2w48D_ |
|
not modelled |
96.5 |
20 |
PDB header:transcription
Chain: D: PDB Molecule:sorbitol operon regulator;
PDBTitle: crystal structure of the full-length sorbitol operon2 regulator sorc from klebsiella pneumoniae
|
| 48 | d2ahua2 |
|
not modelled |
96.5 |
21 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 49 | d2g39a1 |
|
not modelled |
96.5 |
19 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 50 | d3efba1 |
|
not modelled |
96.3 |
20 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 51 | c1w2wJ_ |
|
not modelled |
96.0 |
16 |
PDB header:isomerase
Chain: J: PDB Molecule:5-methylthioribose-1-phosphate isomerase;
PDBTitle: crystal structure of yeast ypr118w, a methylthioribose-1-2 phosphate isomerase related to regulatory eif2b subunits
|
| 52 | c2hj0A_ |
|
not modelled |
95.9 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:putative citrate lyase, alfa subunit;
PDBTitle: crystal structure of the putative alfa subunit of citrate lyase in2 complex with citrate from streptococcus mutans, northeast structural3 genomics target smr12 (casp target).
|
| 53 | d2r5fa1 |
|
not modelled |
95.7 |
22 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 54 | d2okga1 |
|
not modelled |
95.4 |
24 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 55 | c2o0mA_ |
|
not modelled |
95.4 |
18 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator, sorc family;
PDBTitle: the crystal structure of the putative sorc family transcriptional2 regulator from enterococcus faecalis
|
| 56 | d2o0ma1 |
|
not modelled |
95.4 |
18 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:SorC sugar-binding domain-like |
| 57 | c1xr4B_ |
|
not modelled |
95.2 |
21 |
PDB header:hydrolase/transferase
Chain: B: PDB Molecule:putative citrate lyase alpha chain/citrate-acp transferase;
PDBTitle: x-ray crystal structure of putative citrate lyase alpha chain/citrate-2 acp transferase [salmonella typhimurium]
|
| 58 | c3d3uA_ |
|
not modelled |
92.6 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:4-hydroxybutyrate coa-transferase;
PDBTitle: crystal structure of 4-hydroxybutyrate coa-transferase (abft-2) from2 porphyromonas gingivalis. northeast structural genomics consortium3 target pgr26
|
| 59 | c2ri0B_ |
|
not modelled |
91.4 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: crystal structure of glucosamine 6-phosphate deaminase (nagb) from s.2 mutans
|
| 60 | c2nvvF_ |
|
not modelled |
88.7 |
21 |
PDB header:hydrolase
Chain: F: PDB Molecule:acetyl-coa hydrolase/transferase family protein;
PDBTitle: crystal structure of the putative acetyl-coa hydrolase/transferase2 pg1013 from porphyromonas gingivalis, northeast structural genomics3 target pgr16.
|
| 61 | c2g39A_ |
|
not modelled |
88.5 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:acetyl-coa hydrolase;
PDBTitle: crystal structure of coenzyme a transferase from pseudomonas2 aeruginosa
|
| 62 | d2g39a2 |
|
not modelled |
88.3 |
17 |
Fold:NagB/RpiA/CoA transferase-like
Superfamily:NagB/RpiA/CoA transferase-like
Family:CoA transferase alpha subunit-like |
| 63 | c2bkxB_ |
|
not modelled |
79.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:glucosamine-6-phosphate deaminase;
PDBTitle: structure and kinetics of a monomeric glucosamine-6-2 phosphate deaminase: missing link of the nagb superfamily
|
| 64 | c2zbcH_ |
|
not modelled |
73.4 |
14 |
PDB header:transcription
Chain: H: PDB Molecule:83aa long hypothetical transcriptional regulator asnc;
PDBTitle: crystal structure of sts042, a stand-alone ram module protein, from2 hyperthermophilic archaeon sulfolobus tokodaii strain7.
|
| 65 | c2e1cA_ |
|
not modelled |
72.3 |
21 |
PDB header:transcription/dna
Chain: A: PDB Molecule:putative hth-type transcriptional regulator ph1519;
PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
|
| 66 | d2nzca1 |
|
not modelled |
71.7 |
20 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:TM1266-like |
| 67 | c2gqqB_ |
|
not modelled |
70.6 |
10 |
PDB header:transcription
Chain: B: PDB Molecule:leucine-responsive regulatory protein;
PDBTitle: crystal structure of e. coli leucine-responsive regulatory protein2 (lrp)
|
| 68 | c1gpjA_ |
|
not modelled |
66.7 |
16 |
PDB header:reductase
Chain: A: PDB Molecule:glutamyl-trna reductase;
PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
|
| 69 | c3eeyJ_ |
|
not modelled |
62.2 |
13 |
PDB header:transferase
Chain: J: PDB Molecule:putative rrna methylase;
PDBTitle: crystal structure of putative rrna-methylase from clostridium2 thermocellum
|
| 70 | d1i1ga2 |
|
not modelled |
61.1 |
19 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 71 | c1i1gA_ |
|
not modelled |
60.2 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator lrpa;
PDBTitle: crystal structure of the lrp-like transcriptional regulator from the2 archaeon pyrococcus furiosus
|
| 72 | c2cg4B_ |
|
not modelled |
56.9 |
13 |
PDB header:transcription
Chain: B: PDB Molecule:regulatory protein asnc;
PDBTitle: structure of e.coli asnc
|
| 73 | d1iz0a2 |
|
not modelled |
56.9 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 74 | c2g1uA_ |
|
not modelled |
54.4 |
9 |
PDB header:transport protein
Chain: A: PDB Molecule:hypothetical protein tm1088a;
PDBTitle: crystal structure of a putative transport protein (tm1088a) from2 thermotoga maritima at 1.50 a resolution
|
| 75 | c2jsxA_ |
|
not modelled |
53.4 |
23 |
PDB header:chaperone
Chain: A: PDB Molecule:protein napd;
PDBTitle: solution structure of the e. coli tat proofreading2 chaperone protein napd
|
| 76 | c3mw8A_ |
|
not modelled |
53.2 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: crystal structure of an uroporphyrinogen-iii synthase (sama_3255) from2 shewanella amazonensis sb2b at 1.65 a resolution
|
| 77 | c2e1aD_ |
|
not modelled |
51.3 |
14 |
PDB header:transcription
Chain: D: PDB Molecule:75aa long hypothetical regulatory protein asnc;
PDBTitle: crystal structure of ffrp-dm1
|
| 78 | c3lhiA_ |
|
not modelled |
49.8 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative 6-phosphogluconolactonase;
PDBTitle: crystal structure of putative 6-2 phosphogluconolactonase(yp_207848.1) from neisseria3 gonorrhoeae fa 1090 at 1.33 a resolution
|
| 79 | d1wd7a_ |
|
not modelled |
47.8 |
14 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
| 80 | d1jr2a_ |
|
not modelled |
46.7 |
9 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
| 81 | c1jr2A_ |
|
not modelled |
46.7 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: structure of uroporphyrinogen iii synthase
|
| 82 | c2qi2A_ |
|
not modelled |
46.0 |
17 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein pelota related protein;
PDBTitle: crystal structure of the thermoplasma acidophilum pelota2 protein
|
| 83 | c2e7xA_ |
|
not modelled |
42.9 |
7 |
PDB header:transcription regulator
Chain: A: PDB Molecule:150aa long hypothetical transcriptional regulator;
PDBTitle: structure of the lrp/asnc like transcriptional regulator from2 sulfolobus tokodaii 7 complexed with its cognate ligand
|
| 84 | d2cg4a2 |
|
not modelled |
42.2 |
13 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 85 | d1z7ga1 |
|
not modelled |
41.7 |
15 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 86 | c3p9zA_ |
|
not modelled |
41.0 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:uroporphyrinogen iii cosynthase (hemd);
PDBTitle: crystal structure of uroporphyrinogen-iii synthetase from helicobacter2 pylori 26695
|
| 87 | d1cjba_ |
|
not modelled |
40.7 |
19 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 88 | c4a10A_ |
|
not modelled |
40.7 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:octenoyl-coa reductase/carboxylase;
PDBTitle: apo-structure of 2-octenoyl-coa carboxylase reductase cinf from2 streptomyces sp.
|
| 89 | c3dfzB_ |
|
not modelled |
40.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:precorrin-2 dehydrogenase;
PDBTitle: sirc, precorrin-2 dehydrogenase
|
| 90 | c3ibwA_ |
|
not modelled |
39.9 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:gtp pyrophosphokinase;
PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
|
| 91 | d1p17b_ |
|
not modelled |
39.2 |
19 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 92 | d1htwa_ |
|
not modelled |
38.7 |
16 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:YjeE-like |
| 93 | d1ucra_ |
|
not modelled |
37.1 |
20 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Dissimilatory sulfite reductase DsvD |
| 94 | d1p0fa2 |
|
not modelled |
37.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 95 | c3c85A_ |
|
not modelled |
36.4 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:putative glutathione-regulated potassium-efflux system
PDBTitle: crystal structure of trka domain of putative glutathione-regulated2 potassium-efflux kefb from vibrio parahaemolyticus
|
| 96 | d2cfxa2 |
|
not modelled |
35.6 |
13 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 97 | c1y89B_ |
|
not modelled |
33.8 |
27 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:devb protein;
PDBTitle: crystal structure of devb protein
|
| 98 | c2npnA_ |
|
not modelled |
33.6 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:putative cobalamin synthesis related protein;
PDBTitle: crystal structure of putative cobalamin synthesis related protein2 (cobf) from corynebacterium diphtheriae
|
| 99 | c3nd1B_ |
|
not modelled |
33.5 |
8 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-6a synthase/cobf protein;
PDBTitle: crystal structure of precorrin-6a synthase from rhodobacter capsulatus
|
| 100 | c2djwF_ |
|
not modelled |
33.4 |
13 |
PDB header:unknown function
Chain: F: PDB Molecule:probable transcriptional regulator, asnc family;
PDBTitle: crystal structure of ttha0845 from thermus thermophilus hb8
|
| 101 | d1kola2 |
|
not modelled |
33.4 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 102 | d1hgxa_ |
|
not modelled |
33.3 |
19 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 103 | d2cyya2 |
|
not modelled |
33.1 |
21 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:Lrp/AsnC-like transcriptional regulator C-terminal domain |
| 104 | d1wd5a_ |
|
not modelled |
32.7 |
14 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 105 | c2p6tH_ |
|
not modelled |
31.5 |
11 |
PDB header:transcription
Chain: H: PDB Molecule:transcriptional regulator, lrp/asnc family;
PDBTitle: crystal structure of transcriptional regulator nmb0573 and l-leucine2 complex from neisseria meningitidis
|
| 106 | d1gpja2 |
|
not modelled |
30.8 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Aminoacid dehydrogenase-like, C-terminal domain |
| 107 | d1ykfa2 |
|
not modelled |
30.7 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 108 | c2vbzA_ |
|
not modelled |
29.8 |
11 |
PDB header:dna-binding protein
Chain: A: PDB Molecule:transcriptional regulatory protein;
PDBTitle: feast or famine regulatory protein (rv3291c)from m.2 tuberculosis complexed with l-tryptophan
|
| 109 | d1i5ea_ |
|
not modelled |
29.5 |
26 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 110 | d1cdoa2 |
|
not modelled |
29.5 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 111 | c2jbvA_ |
|
not modelled |
29.4 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:choline oxidase;
PDBTitle: crystal structure of choline oxidase reveals insights into2 the catalytic mechanism
|
| 112 | c3d4oA_ |
|
not modelled |
29.3 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
| 113 | d1e3ia2 |
|
not modelled |
28.9 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Alcohol dehydrogenase-like, C-terminal domain |
| 114 | c3fwzA_ |
|
not modelled |
28.9 |
15 |
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein ybal;
PDBTitle: crystal structure of trka-n domain of inner membrane protein ybal from2 escherichia coli
|
| 115 | d1fmta2 |
|
not modelled |
28.6 |
25 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 116 | d2hmva1 |
|
not modelled |
28.5 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
| 117 | c3rfoA_ |
|
not modelled |
28.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: crystal structure of methyionyl-trna formyltransferase from bacillus2 anthracis
|
| 118 | d1tc1a_ |
|
not modelled |
28.1 |
19 |
Fold:PRTase-like
Superfamily:PRTase-like
Family:Phosphoribosyltransferases (PRTases) |
| 119 | d2bw0a2 |
|
not modelled |
28.0 |
16 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
| 120 | c1yfzA_ |
|
not modelled |
27.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:hypoxanthine-guanine phosphoribosyltransferase;
PDBTitle: novel imp binding in feedback inhibition of hypoxanthine-guanine2 phosphoribosyltransferase from thermoanaerobacter tengcongensis
|