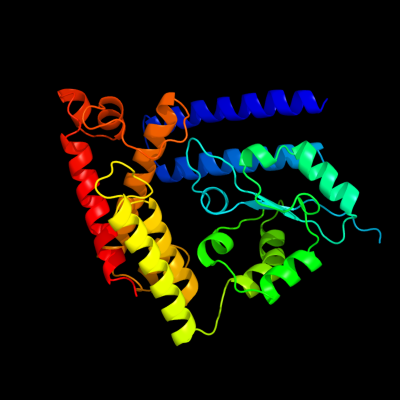
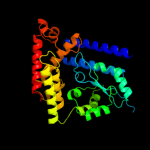
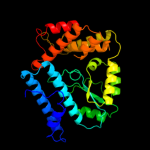
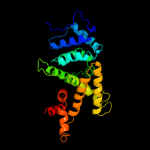
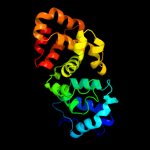
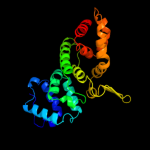
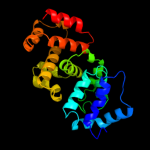
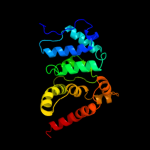
1 c1v4aA_
100.0
15
PDB header: transferaseChain: A: PDB Molecule: glutamate-ammonia-ligase adenylyltransferase;PDBTitle: structure of the n-terminal domain of escherichia coli2 glutamine synthetase adenylyltransferase
2 c3k7dA_
100.0
17
PDB header: transferaseChain: A: PDB Molecule: glutamate-ammonia-ligase adenylyltransferase;PDBTitle: c-terminal (adenylylation) domain of e.coli glutamine synthetase2 adenylyltransferase
3 c3aqnA_
100.0
12
PDB header: transferaseChain: A: PDB Molecule: poly(a) polymerase;PDBTitle: complex structure of bacterial protein (apo form ii)
4 d1v4aa2
100.0
14
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: GlnE-like domain5 d1v4aa1
99.9
18
Fold: Four-helical up-and-down bundleSuperfamily: Nucleotidyltransferase substrate binding subunit/domainFamily: Glutamine synthase adenylyltransferase GlnE, domain 26 c1u8sB_
99.8
14
PDB header: transcriptionChain: B: PDB Molecule: glycine cleavage system transcriptionalPDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
7 c2nyiB_
99.8
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: unknown protein;PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
8 c1miyB_
99.7
16
PDB header: translation, transferaseChain: B: PDB Molecule: trna cca-adding enzyme;PDBTitle: crystal structure of bacillus stearothermophilus cca-adding enzyme in2 complex with ctp
9 c1vfgB_
99.7
16
PDB header: transferase/rnaChain: B: PDB Molecule: poly a polymerase;PDBTitle: crystal structure of trna nucleotidyltransferase complexed2 with a primer trna and an incoming atp analog
10 d1vfga1
99.7
15
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like11 d1miwa1
99.6
15
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like12 c3h37B_
99.5
14
PDB header: transferaseChain: B: PDB Molecule: trna nucleotidyl transferase-related protein;PDBTitle: the structure of cca-adding enzyme apo form i
13 c1ou5A_
99.5
13
PDB header: translation, transferaseChain: A: PDB Molecule: trna cca-adding enzyme;PDBTitle: crystal structure of human cca-adding enzyme
14 d1ou5a1
99.3
13
Fold: Poly A polymerase C-terminal region-likeSuperfamily: Poly A polymerase C-terminal region-likeFamily: Poly A polymerase C-terminal region-like15 d2pq7a1
98.9
25
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain16 d2qgsa1
98.7
22
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain17 d3djba1
98.6
26
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain18 c2f06B_
98.5
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
19 c3ibwA_
98.3
11
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
20 d1knya2
98.2
14
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Kanamycin nucleotidyltransferase (KNTase), N-terminal domain21 d1u8sa2
not modelled
98.1
15
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor22 d3dtoa1
not modelled
98.1
29
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain23 d3b57a1
not modelled
98.0
21
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain24 d1zpva1
not modelled
98.0
18
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: SP0238-like25 d1u8sa1
not modelled
98.0
18
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor26 c3gw7A_
not modelled
97.9
20
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein yedj;PDBTitle: crystal structure of a metal-dependent phosphohydrolase2 with conserved hd domain (yedj) from escherichia coli in3 complex with nickel ions. northeast structural genomics4 consortium target er63
27 c3p96A_
not modelled
97.9
12
PDB header: hydrolaseChain: A: PDB Molecule: phosphoserine phosphatase serb;PDBTitle: crystal structure of phosphoserine phosphatase serb from mycobacterium2 avium, native form
28 c3m5fA_
not modelled
97.9
20
PDB header: hydrolaseChain: A: PDB Molecule: metal dependent phosphohydrolase;PDBTitle: structure of mj0384, a cas3 protein from methanocaldococcus jannaschii
29 c2q14A_
not modelled
97.9
18
PDB header: hydrolaseChain: A: PDB Molecule: phosphohydrolase;PDBTitle: crystal structure of phosphohydrolase (bt4208) from bacteroides2 thetaiotaomicron vpi-5482 at 2.20 a resolution
30 c2o08B_
not modelled
97.9
26
PDB header: hydrolaseChain: B: PDB Molecule: bh1327 protein;PDBTitle: crystal structure of a putative hd superfamily hydrolase (bh1327) from2 bacillus halodurans at 1.90 a resolution
31 c3ccgA_
97.8
22
PDB header: hydrolaseChain: A: PDB Molecule: hd superfamily hydrolase;PDBTitle: crystal structure of predicted hd superfamily hydrolase involved in2 nad metabolism (np_347894.1) from clostridium acetobutylicum at 1.503 a resolution
32 c3skdA_
not modelled
97.8
15
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein tthb187;PDBTitle: crystal structure of the thermus thermophilus cas3 hd domain in the2 presence of ni2+
33 c2ogiA_
not modelled
97.6
25
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein sag1661;PDBTitle: crystal structure of a putative metal dependent phosphohydrolase2 (sag1661) from streptococcus agalactiae serogroup v at 1.85 a3 resolution
34 d2pjqa1
not modelled
97.5
18
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain35 d1u6za1
not modelled
97.5
25
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: Ppx associated domain36 c3n0vD_
not modelled
97.5
8
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
37 c3o1lB_
not modelled
97.5
15
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
38 d2o6ia1
not modelled
97.4
15
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain39 c2o6iA_
not modelled
97.4
15
PDB header: hydrolaseChain: A: PDB Molecule: hd domain protein;PDBTitle: structure of an enterococcus faecalis hd domain phosphohydrolase
40 d1ygya3
not modelled
97.4
19
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain41 c3obiC_
not modelled
97.3
15
PDB header: hydrolaseChain: C: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
42 c2dqbB_
not modelled
97.3
20
PDB header: hydrolase, dna binding proteinChain: B: PDB Molecule: deoxyguanosinetriphosphate triphosphohydrolase, putative;PDBTitle: crystal structure of dntp triphosphohydrolase from thermus2 thermophilus hb8, which is homologous to dgtp triphosphohydrolase
43 c3nrbD_
not modelled
97.2
17
PDB header: hydrolaseChain: D: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
44 d2heka1
not modelled
97.2
16
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain45 c1knyA_
not modelled
97.2
19
PDB header: transferaseChain: A: PDB Molecule: kanamycin nucleotidyltransferase;PDBTitle: kanamycin nucleotidyltransferase
46 c2rffA_
not modelled
97.1
18
PDB header: transferaseChain: A: PDB Molecule: putative nucleotidyltransferase;PDBTitle: crystal structure of a putative nucleotidyltransferase2 (np_343093.1) from sulfolobus solfataricus at 1.40 a3 resolution
47 c3hc1A_
not modelled
97.0
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized hdod domain protein;PDBTitle: crystal structure of hdod domain protein with unknown function2 (np_953345.1) from geobacter sulfurreducens at 1.90 a resolution
48 d1sc6a3
not modelled
97.0
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain49 d2f1fa1
not modelled
96.9
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like50 c3l76B_
96.9
20
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of aspartate kinase from synechocystis
51 c2floA_
not modelled
96.9
20
PDB header: hydrolaseChain: A: PDB Molecule: exopolyphosphatase;PDBTitle: crystal structure of exopolyphosphatase (ppx) from e. coli o157:h7
52 d2pc6a2
not modelled
96.8
15
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like53 d2gz4a1
not modelled
96.8
18
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain54 c3m1tA_
not modelled
96.8
15
PDB header: hydrolaseChain: A: PDB Molecule: putative phosphohydrolase;PDBTitle: crystal structure of putative phosphohydrolase (yp_929327.1) from2 shewanella amazonensis sb2b at 1.62 a resolution
55 c3louB_
not modelled
96.8
16
PDB header: hydrolaseChain: B: PDB Molecule: formyltetrahydrofolate deformylase;PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
56 c3hi0B_
not modelled
96.7
11
PDB header: hydrolaseChain: B: PDB Molecule: putative exopolyphosphatase;PDBTitle: crystal structure of putative exopolyphosphatase (17739545) from2 agrobacterium tumefaciens str. c58 (dupont) at 2.30 a resolution
57 c3memA_
not modelled
96.7
17
PDB header: signaling proteinChain: A: PDB Molecule: putative signal transduction protein;PDBTitle: crystal structure of a putative signal transduction protein2 (maqu_0641) from marinobacter aquaeolei vt8 at 2.25 a resolution
58 d2f06a1
not modelled
96.7
13
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like59 c1ygyA_
not modelled
96.7
14
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
60 c3u1nC_
not modelled
96.6
20
PDB header: hydrolaseChain: C: PDB Molecule: sam domain and hd domain-containing protein 1;PDBTitle: structure of the catalytic core of human samhd1
61 d1vqra_
not modelled
96.6
13
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: modified HD domain62 d1no5a_
not modelled
96.6
21
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase63 c2f1fA_
not modelled
96.4
14
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase isozyme iii small subunit;PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
64 d1ylqa1
not modelled
96.4
19
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase65 d1taza_
not modelled
96.2
18
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: PDEase66 c2pc6C_
not modelled
96.1
15
PDB header: lyaseChain: C: PDB Molecule: probable acetolactate synthase isozyme iii (small subunit);PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
67 d2fgca2
not modelled
96.1
12
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: IlvH-like68 c3ljvA_
not modelled
95.8
10
PDB header: transcriptionChain: A: PDB Molecule: mmoq response regulator;PDBTitle: crystal structure of mmoq response regulator (fragment 29-302) from2 methylococcus capsulatus str. bath, northeast structural genomics3 consortium target mcr175m
69 c2fgcA_
not modelled
95.8
12
PDB header: transferaseChain: A: PDB Molecule: acetolactate synthase, small subunit;PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
70 d2f06a2
not modelled
95.8
20
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: BT0572-like71 c3i7aA_
not modelled
95.7
7
PDB header: hydrolaseChain: A: PDB Molecule: putative metal-dependent phosphohydrolase;PDBTitle: crystal structure of putative metal-dependent phosphohydrolase2 (yp_926882.1) from shewanella amazonensis sb2b at 2.06 a resolution
72 c2pgsA_
not modelled
95.7
18
PDB header: hydrolaseChain: A: PDB Molecule: putative deoxyguanosinetriphosphate triphosphohydrolase;PDBTitle: crystal structure of a putative deoxyguanosinetriphosphate2 triphosphohydrolase from pseudomonas syringae pv. phaseolicola 1448a
73 c1zklA_
not modelled
95.6
17
PDB header: hydrolaseChain: A: PDB Molecule: high-affinity camp-specific 3',5'-cyclicPDBTitle: multiple determinants for inhibitor selectivity of cyclic2 nucleotide phosphodiesterases
74 d1wota_
not modelled
95.5
26
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase75 d1so2a_
not modelled
95.5
19
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: PDEase76 d1f0ja_
not modelled
95.4
20
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: PDEase77 c3g3nA_
not modelled
95.2
17
PDB header: hydrolaseChain: A: PDB Molecule: high affinity camp-specific 3',5'-cyclicPDBTitle: pde7a catalytic domain in complex with 3-(2,6-2 difluorophenyl)-2-(methylthio)quinazolin-4(3h)-one
78 c2zhoB_
not modelled
95.1
19
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of the regulatory subunit of aspartate2 kinase from thermus thermophilus (ligand free form)
79 c3qi4A_
not modelled
94.8
15
PDB header: hydrolaseChain: A: PDB Molecule: high affinity cgmp-specific 3',5'-cyclic phosphodiesterasePDBTitle: crystal structure of pde9a(q453e) in complex with ibmx
80 c2dtjA_
not modelled
94.8
17
PDB header: transferaseChain: A: PDB Molecule: aspartokinase;PDBTitle: crystal structure of regulatory subunit of aspartate kinase2 from corynebacterium glutamicum
81 c3kq5A_
not modelled
94.4
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical cytosolic protein;PDBTitle: crystal structure of an uncharacterized protein from coxiella burnetii
82 d1phza1
not modelled
93.7
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain83 c3c66B_
not modelled
93.7
18
PDB header: transferaseChain: B: PDB Molecule: poly(a) polymerase;PDBTitle: yeast poly(a) polymerase in complex with fip1 residues 80-105
84 c1xotB_
not modelled
93.3
19
PDB header: hydrolaseChain: B: PDB Molecule: camp-specific 3',5'-cyclic phosphodiesterase 4b;PDBTitle: catalytic domain of human phosphodiesterase 4b in complex with2 vardenafil
85 d2qmwa2
not modelled
93.1
25
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Phenylalanine metabolism regulatory domain86 c2r8qA_
not modelled
93.0
15
PDB header: hydrolaseChain: A: PDB Molecule: class i phosphodiesterase pdeb1;PDBTitle: structure of lmjpdeb1 in complex with ibmx
87 c3luyA_
not modelled
92.8
17
PDB header: isomeraseChain: A: PDB Molecule: probable chorismate mutase;PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
88 c3mwbA_
not modelled
92.8
19
PDB header: lyaseChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
89 c3bg2A_
not modelled
92.8
23
PDB header: hydrolaseChain: A: PDB Molecule: dgtp triphosphohydrolase;PDBTitle: crystal structure of deoxyguanosinetriphosphate triphosphohydrolase2 from flavobacterium sp. med217
90 c3mtjA_
not modelled
92.7
22
PDB header: oxidoreductaseChain: A: PDB Molecule: homoserine dehydrogenase;PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
91 d1q79a2
not modelled
92.4
11
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Poly(A) polymerase, PAP, N-terminal domain92 c2qmxB_
not modelled
92.3
15
PDB header: ligaseChain: B: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of l-phe inhibited prephenate dehydratase from2 chlorobium tepidum tls
93 c2ounA_
not modelled
92.2
19
PDB header: hydrolaseChain: A: PDB Molecule: camp and camp-inhibited cgmp 3',5'-cyclicPDBTitle: crystal structure of pde10a2 in complex with amp
94 d1jmsa4
not modelled
92.1
24
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like95 c1q78A_
not modelled
92.0
11
PDB header: transferaseChain: A: PDB Molecule: poly(a) polymerase alpha;PDBTitle: crystal structure of poly(a) polymerase in complex with 3'-2 datp and magnesium chloride
96 c1xozA_
not modelled
91.9
16
PDB header: hydrolaseChain: A: PDB Molecule: cgmp-specific 3',5'-cyclic phosphodiesterase;PDBTitle: catalytic domain of human phosphodiesterase 5a in complex2 with tadalafil
97 c3ibjB_
not modelled
91.8
14
PDB header: hydrolaseChain: B: PDB Molecule: cgmp-dependent 3',5'-cyclic phosphodiesterase;PDBTitle: x-ray structure of pde2a
98 d1y2ka1
not modelled
91.5
15
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: PDEase99 d1tbfa_
not modelled
91.1
15
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: PDEase100 d2q66a2
not modelled
91.1
21
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Poly(A) polymerase, PAP, N-terminal domain101 c1z1lA_
not modelled
90.6
14
PDB header: hydrolaseChain: A: PDB Molecule: cgmp-dependent 3',5'-cyclic phosphodiesterase;PDBTitle: the crystal structure of the phosphodiesterase 2a catalytic2 domain
102 d3dy8a1
not modelled
90.5
15
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: PDEase103 c1kdhA_
not modelled
90.3
16
PDB header: transferase/dnaChain: A: PDB Molecule: terminal deoxynucleotidyltransferase shortPDBTitle: binary complex of murine terminal deoxynucleotidyl2 transferase with a primer single stranded dna
104 c2re1A_
not modelled
90.0
19
PDB header: transferaseChain: A: PDB Molecule: aspartokinase, alpha and beta subunits;PDBTitle: crystal structure of aspartokinase alpha and beta subunits
105 c3mzoA_
not modelled
89.7
20
PDB header: hydrolaseChain: A: PDB Molecule: lin2634 protein;PDBTitle: crystal structure of a hd-domain phosphohydrolase (lin2634) from2 listeria innocua at 1.98 a resolution
106 c3nqwB_
not modelled
88.6
19
PDB header: hydrolaseChain: B: PDB Molecule: cg11900;PDBTitle: a metazoan ortholog of spot hydrolyzes ppgpp and plays a role in2 starvation responses
107 c2o8hA_
not modelled
88.3
18
PDB header: hydrolaseChain: A: PDB Molecule: phosphodiesterase-10a;PDBTitle: crystal structure of the catalytic domain of rat2 phosphodiesterase 10a
108 c2cqzA_
not modelled
88.2
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 177aa long hypothetical protein;PDBTitle: crystal structure of ph0347 protein from pyrococcus horikoshii ot3
109 d2h44a1
not modelled
87.4
15
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: PDEase110 c3jz0B_
not modelled
87.2
23
PDB header: transferase/antibioticChain: B: PDB Molecule: lincosamide nucleotidyltransferase;PDBTitle: linb complexed with clindamycin and ampcpp
111 c3bjcA_
not modelled
87.0
15
PDB header: hydrolaseChain: A: PDB Molecule: cgmp-specific 3',5'-cyclic phosphodiesterase;PDBTitle: crystal structure of the pde5a catalytic domain in complex2 with a novel inhibitor
112 c2huoA_
not modelled
85.6
20
PDB header: oxidoreductaseChain: A: PDB Molecule: inositol oxygenase;PDBTitle: crystal structure of mouse myo-inositol oxygenase in complex with2 substrate
113 d3bxda1
not modelled
85.6
20
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: MioX-like114 d2hmfa3
not modelled
85.1
14
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Aspartokinase allosteric domain-like115 d1ynba1
not modelled
84.6
24
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain116 c1sz1A_
not modelled
83.2
19
PDB header: transferase/rnaChain: A: PDB Molecule: trna nucleotidyltransferase;PDBTitle: mechanism of cca-adding enzymes specificity revealed by crystal2 structures of ternary complexes
117 d2ibna1
not modelled
83.0
23
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: MioX-like118 d2paqa1
not modelled
81.8
24
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain119 c2qmwA_
not modelled
81.6
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: prephenate dehydratase;PDBTitle: the crystal structure of the prephenate dehydratase (pdt) from2 staphylococcus aureus subsp. aureus mu50
120 d2fmpa3
not modelled
80.6
13
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: DNA polymerase beta-like