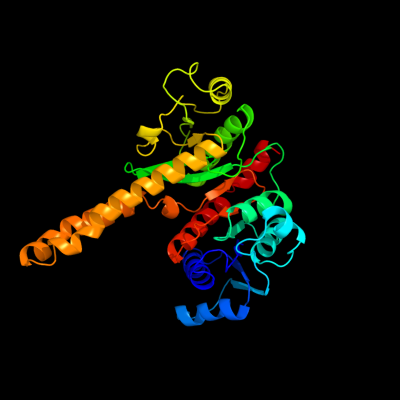
| 1 | d1u7na_
|
|
 |
100.0 |
37 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PlsX-like |
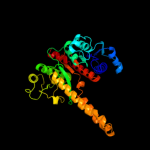
| 2 | d1vi1a_
|
|
 |
100.0 |
36 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PlsX-like |
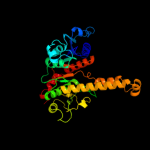
| 3 | d1xcoa_
|
|
 |
100.0 |
22 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 4 | d2af4c1
|
|
 |
100.0 |
19 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 5 | d1r5ja_
|
|
 |
100.0 |
22 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 6 | c1ycoA_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:branched-chain phosphotransacylase;
PDBTitle: crystal structure of a branched-chain phosphotransacylase from2 enterococcus faecalis v583
|
| 7 | c3tngA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:lmo1369 protein;
PDBTitle: the crystal structure of a possible phosphate acetyl/butaryl2 transferase from listeria monocytogenes egd-e.
|
| 8 | d1vmia_
|
|
 |
100.0 |
19 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Phosphotransacetylase |
| 9 | c1vmiA_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:putative phosphate acetyltransferase;
PDBTitle: crystal structure of putative phosphate acetyltransferase2 (np_416953.1) from escherichia coli k12 at 2.32 a resolution
|
| 10 | d1ptma_
|
|
 |
100.0 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
| 11 | c1yxoB_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 1;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic protein pdxa2 pa0593
|
| 12 | d1r8ka_
|
|
 |
100.0 |
17 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
| 13 | c2hi1A_
|
|
 |
98.4 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 2;
PDBTitle: the structure of a putative 4-hydroxythreonine-4-phosphate2 dehydrogenase from salmonella typhimurium.
|
| 14 | d1qgoa_
|
|
 |
74.7 |
12 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Cobalt chelatase CbiK |
| 15 | d1gmla_
|
|
 |
64.9 |
19 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:Group II chaperonin (CCT, TRIC), apical domain |
| 16 | c2phjA_
|
|
 |
56.6 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-nucleotidase sure;
PDBTitle: crystal structure of sure protein from aquifex aeolicus
|
| 17 | c3qd5B_
|
|
 |
56.0 |
23 |
PDB header:isomerase
Chain: B: PDB Molecule:putative ribose-5-phosphate isomerase;
PDBTitle: crystal structure of a putative ribose-5-phosphate isomerase from2 coccidioides immitis solved by combined iodide ion sad and mr
|
| 18 | c3lyhB_
|
|
 |
55.8 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:cobalamin (vitamin b12) biosynthesis cbix protein;
PDBTitle: crystal structure of putative cobalamin (vitamin b12) biosynthesis2 cbix protein (yp_958415.1) from marinobacter aquaeolei vt8 at 1.60 a3 resolution
|
| 19 | c3gdwA_
|
|
 |
53.7 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:sigma-54 interaction domain protein;
PDBTitle: crystal structure of sigma-54 interaction domain protein from2 enterococcus faecalis
|
| 20 | d1tjna_
|
|
 |
52.2 |
23 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:CbiX-like |
| 21 | c1tjnA_ |
|
not modelled |
52.2 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:sirohydrochlorin cobaltochelatase;
PDBTitle: crystal structure of hypothetical protein af0721 from archaeoglobus2 fulgidus
|
| 22 | c2pjuD_ |
|
not modelled |
52.0 |
21 |
PDB header:transcription
Chain: D: PDB Molecule:propionate catabolism operon regulatory protein;
PDBTitle: crystal structure of propionate catabolism operon2 regulatory protein prpr
|
| 23 | d1kzyc2 |
|
not modelled |
48.1 |
29 |
Fold:BRCT domain
Superfamily:BRCT domain
Family:53BP1 |
| 24 | c2ja9A_ |
|
not modelled |
46.9 |
14 |
PDB header:rna-binding protein
Chain: A: PDB Molecule:exosome complex exonuclease rrp40;
PDBTitle: structure of the n-terminal deletion of yeast exosome2 component rrp40
|
| 25 | d1q3qa3 |
|
not modelled |
43.6 |
19 |
Fold:GroEL-intermediate domain like
Superfamily:GroEL-intermediate domain like
Family:Group II chaperonin (CCT, TRIC), intermediate domain |
| 26 | d1j9ja_ |
|
not modelled |
43.5 |
16 |
Fold:SurE-like
Superfamily:SurE-like
Family:SurE-like |
| 27 | d1wu2a3 |
|
not modelled |
42.9 |
33 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 28 | c2v4oB_ |
|
not modelled |
42.2 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:multifunctional protein sur e;
PDBTitle: crystal structure of salmonella typhimurium sure at 2.752 angstrom resolution in monoclinic form
|
| 29 | c2xvzA_ |
|
not modelled |
40.6 |
19 |
PDB header:metal binding protein
Chain: A: PDB Molecule:chelatase, putative;
PDBTitle: cobalt chelatase cbik (periplasmatic) from desulvobrio2 vulgaris hildenborough (co-crystallized with cobalt)
|
| 30 | d1y5ea1 |
|
not modelled |
39.5 |
16 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 31 | d2pjua1 |
|
not modelled |
38.5 |
22 |
Fold:Chelatase-like
Superfamily:PrpR receptor domain-like
Family:PrpR receptor domain-like |
| 32 | d1fuia2 |
|
not modelled |
38.3 |
14 |
Fold:FucI/AraA N-terminal and middle domains
Superfamily:FucI/AraA N-terminal and middle domains
Family:L-fucose isomerase, N-terminal and second domains |
| 33 | d2ja9a2 |
|
not modelled |
38.2 |
15 |
Fold:Eukaryotic type KH-domain (KH-domain type I)
Superfamily:Eukaryotic type KH-domain (KH-domain type I)
Family:Eukaryotic type KH-domain (KH-domain type I) |
| 34 | d2ftsa3 |
|
not modelled |
36.8 |
45 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 35 | c3gx1A_ |
|
not modelled |
35.4 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin1832 protein;
PDBTitle: crystal structure of a domain of lin1832 from listeria innocua
|
| 36 | d3ct6a1 |
|
not modelled |
35.2 |
29 |
Fold:PTS system fructose IIA component-like
Superfamily:PTS system fructose IIA component-like
Family:DhaM-like |
| 37 | d1uz5a3 |
|
not modelled |
34.8 |
45 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 38 | c3k93A_ |
|
not modelled |
31.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:phage related exonuclease;
PDBTitle: crystal structure of phage related exonuclease (yp_719632.1) from2 haemophilus somnus 129pt at 2.15 a resolution
|
| 39 | d3b48a1 |
|
not modelled |
30.1 |
24 |
Fold:PTS system fructose IIA component-like
Superfamily:PTS system fructose IIA component-like
Family:DhaM-like |
| 40 | c3imkA_ |
|
not modelled |
29.7 |
22 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative molybdenum carrier protein;
PDBTitle: crystal structure of putative molybdenum carrier protein (yp_461806.1)2 from syntrophus aciditrophicus sb at 1.45 a resolution
|
| 41 | d1pkpa1 |
|
not modelled |
28.9 |
19 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 42 | d2uube1 |
|
not modelled |
28.3 |
24 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 43 | d1iowa1 |
|
not modelled |
28.0 |
21 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:D-Alanine ligase N-terminal domain |
| 44 | c3se7A_ |
|
not modelled |
26.7 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:vana;
PDBTitle: ancient vana
|
| 45 | c3lkzB_ |
|
not modelled |
24.6 |
23 |
PDB header:viral protein
Chain: B: PDB Molecule:non-structural protein 5;
PDBTitle: structural and functional analyses of a conserved hydrophobic pocket2 of flavivirus methyltransferase
|
| 46 | d1jlja_ |
|
not modelled |
22.9 |
21 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
| 47 | d1su1a_ |
|
not modelled |
21.8 |
14 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 48 | c1su1A_ |
|
not modelled |
21.8 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yfce;
PDBTitle: structural and biochemical characterization of yfce, a2 phosphoesterase from e. coli
|
| 49 | c1p6gE_ |
|
not modelled |
21.6 |
15 |
PDB header:ribosome
Chain: E: PDB Molecule:30s ribosomal protein s5;
PDBTitle: real space refined coordinates of the 30s subunit fitted2 into the low resolution cryo-em map of the ef-g.gtp state3 of e. coli 70s ribosome
|
| 50 | c1jqsB_ |
|
not modelled |
21.3 |
50 |
PDB header:ribosome
Chain: B: PDB Molecule:elongation factor g;
PDBTitle: fitting of l11 protein and elongation factor g (domain g'2 and v) in the cryo-em map of e. coli 70s ribosome bound3 with ef-g and gmppcp, a nonhydrolysable gtp analog
|
| 51 | d2qale1 |
|
not modelled |
21.2 |
19 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:Translational machinery components |
| 52 | c3i12A_ |
|
not modelled |
20.4 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:d-alanine-d-alanine ligase a;
PDBTitle: the crystal structure of the d-alanyl-alanine synthetase a from2 salmonella enterica subsp. enterica serovar typhimurium str. lt2
|
| 53 | d1v4va_ |
|
not modelled |
19.9 |
21 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
| 54 | d1a6db2 |
|
not modelled |
19.8 |
14 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:Group II chaperonin (CCT, TRIC), apical domain |
| 55 | d1assa_ |
|
not modelled |
17.5 |
17 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:GroEL apical domain-like
Family:Group II chaperonin (CCT, TRIC), apical domain |
| 56 | d1iqoa_ |
|
not modelled |
17.2 |
14 |
Fold:Hypothetical protein MTH1880
Superfamily:Hypothetical protein MTH1880
Family:Hypothetical protein MTH1880 |
| 57 | c2xcuC_ |
|
not modelled |
17.2 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:3-deoxy-d-manno-2-octulosonic acid transferase;
PDBTitle: membrane-embedded monofunctional glycosyltransferase waaa of aquifex2 aeolicus, comlex with cmp
|
| 58 | c2xnkA_ |
|
not modelled |
16.3 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:dna topoisomerase 2-binding protein 1;
PDBTitle: structure and function of the rad9-binding region of the dna damage2 checkpoint adaptor topbp1
|
| 59 | c2e6gI_ |
|
not modelled |
16.1 |
19 |
PDB header:hydrolase
Chain: I: PDB Molecule:5'-nucleotidase sure;
PDBTitle: crystal structure of the stationary phase survival protein sure from2 thermus thermophilus hb8 in complex with phosphate
|
| 60 | c3qfnA_ |
|
not modelled |
16.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of streptococcal asymmetric ap4a hydrolase and2 phosphodiesterase spr1479/saph in complex with inorganic phosphate
|
| 61 | c3af5A_ |
|
not modelled |
15.7 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein ph1404;
PDBTitle: the crystal structure of an archaeal cpsf subunit, ph1404 from2 pyrococcus horikoshii
|
| 62 | d1tqya1 |
|
not modelled |
14.5 |
14 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
| 63 | c2nqqA_ |
|
not modelled |
14.0 |
50 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis protein moea;
PDBTitle: moea r137q
|
| 64 | c3dcjA_ |
|
not modelled |
13.6 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:probable 5'-phosphoribosylglycinamide
PDBTitle: crystal structure of glycinamide formyltransferase (purn)2 from mycobacterium tuberculosis in complex with 5-methyl-5,3 6,7,8-tetrahydrofolic acid derivative
|
| 65 | c2xzmE_ |
|
not modelled |
13.4 |
20 |
PDB header:ribosome
Chain: E: PDB Molecule:ribosomal protein s5 containing protein;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
|
| 66 | c3lfhF_ |
|
not modelled |
13.0 |
19 |
PDB header:transferase
Chain: F: PDB Molecule:phosphotransferase system, mannose/fructose-specific
PDBTitle: crystal structure of manxa from thermoanaerobacter tengcongensis
|
| 67 | c2fu3A_ |
|
not modelled |
13.0 |
27 |
PDB header:biosynthetic protein/structural protein
Chain: A: PDB Molecule:gephyrin;
PDBTitle: crystal structure of gephyrin e-domain
|
| 68 | c2q5cA_ |
|
not modelled |
12.9 |
18 |
PDB header:transcription
Chain: A: PDB Molecule:ntrc family transcriptional regulator;
PDBTitle: crystal structure of ntrc family transcriptional regulator from2 clostridium acetobutylicum
|
| 69 | d1b77a2 |
|
not modelled |
12.5 |
19 |
Fold:DNA clamp
Superfamily:DNA clamp
Family:DNA polymerase processivity factor |
| 70 | c3iprC_ |
|
not modelled |
12.5 |
35 |
PDB header:transferase
Chain: C: PDB Molecule:pts system, iia component;
PDBTitle: crystal structure of the enterococcus faecalis gluconate2 specific eiia phosphotransferase system component
|
| 71 | c3oryA_ |
|
not modelled |
12.4 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:flap endonuclease 1;
PDBTitle: crystal structure of flap endonuclease 1 from hyperthermophilic2 archaeon desulfurococcus amylolyticus
|
| 72 | c2ow8f_ |
|
not modelled |
12.4 |
28 |
PDB header:ribosome
Chain: F: PDB Molecule:
PDBTitle: crystal structure of a 70s ribosome-trna complex reveals functional2 interactions and rearrangements. this file, 2ow8, contains the 30s3 ribosome subunit, two trna, and mrna molecules. 50s ribosome subunit4 is in the file 1vsa.
|
| 73 | d1lqta1 |
|
not modelled |
12.1 |
9 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:C-terminal domain of adrenodoxin reductase-like |
| 74 | d2nn6g3 |
|
not modelled |
11.5 |
13 |
Fold:Eukaryotic type KH-domain (KH-domain type I)
Superfamily:Eukaryotic type KH-domain (KH-domain type I)
Family:Eukaryotic type KH-domain (KH-domain type I) |
| 75 | d1kfia2 |
|
not modelled |
11.3 |
25 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
| 76 | c3bbnE_ |
|
not modelled |
10.6 |
31 |
PDB header:ribosome
Chain: E: PDB Molecule:ribosomal protein s5;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 77 | d1oaoa_ |
|
not modelled |
10.5 |
27 |
Fold:Prismane protein-like
Superfamily:Prismane protein-like
Family:Carbon monoxide dehydrogenase |
| 78 | c2yw3E_ |
|
not modelled |
10.5 |
20 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-
PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
|
| 79 | d1o6ca_ |
|
not modelled |
10.4 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDP-N-acetylglucosamine 2-epimerase |
| 80 | c2jfsA_ |
|
not modelled |
10.4 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:ser-thr phosphatase mspp;
PDBTitle: crystal structure of the ppm ser-thr phosphatase mspp from2 mycobacterium smegmatis in complex with cacodylate
|
| 81 | d2oqea3 |
|
not modelled |
10.4 |
15 |
Fold:Cystatin-like
Superfamily:Amine oxidase N-terminal region
Family:Amine oxidase N-terminal region |
| 82 | c1eg0B_ |
|
not modelled |
10.2 |
22 |
PDB header:ribosome
PDB COMPND:
|
| 83 | d1e4ea1 |
|
not modelled |
10.2 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:D-Alanine ligase N-terminal domain |
| 84 | d1o1xa_ |
|
not modelled |
10.0 |
11 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
| 85 | c2p3qA_ |
|
not modelled |
9.9 |
22 |
PDB header:viral protein,transferase
Chain: A: PDB Molecule:type ii methyltransferase;
PDBTitle: crystal structure of dengue methyltransferase in complex with gpppg2 and s-adenosyl-l-homocysteine
|
| 86 | d2bfwa1 |
|
not modelled |
9.9 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Glycosyl transferases group 1 |
| 87 | c3hp7A_ |
|
not modelled |
9.9 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hemolysin, putative;
PDBTitle: putative hemolysin from streptococcus thermophilus.
|
| 88 | c1uz5A_ |
|
not modelled |
9.6 |
30 |
PDB header:molybdopterin biosynthesis
Chain: A: PDB Molecule:402aa long hypothetical molybdopterin
PDBTitle: the crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikosii
|
| 89 | c2kpmA_ |
|
not modelled |
9.6 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of uncharacterized protein from gene2 locus ne0665 of nitrosomonas europaea. northeast structural3 genomics target ner103a
|
| 90 | c1wu2B_ |
|
not modelled |
9.6 |
31 |
PDB header:structural genomics,biosynthetic protein
Chain: B: PDB Molecule:molybdopterin biosynthesis moea protein;
PDBTitle: crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikoshii ot3
|
| 91 | c2jh3C_ |
|
not modelled |
9.5 |
21 |
PDB header:ribosomal protein
Chain: C: PDB Molecule:ribosomal protein s2-related protein;
PDBTitle: the crystal structure of dr2241 from deinococcus2 radiodurans at 1.9 a resolution reveals a multi-domain3 protein with structural similarity to chelatases but also4 with two additional novel domains
|
| 92 | c1b43A_ |
|
not modelled |
9.3 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:protein (fen-1);
PDBTitle: fen-1 from p. furiosus
|
| 93 | c2wa1A_ |
|
not modelled |
9.2 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:non-structural protein 5;
PDBTitle: structure of the methyltransferase domain from modoc virus,2 a flavivirus with no known vector (nkv)
|
| 94 | d1xo1a1 |
|
not modelled |
9.1 |
10 |
Fold:SAM domain-like
Superfamily:5' to 3' exonuclease, C-terminal subdomain
Family:5' to 3' exonuclease, C-terminal subdomain |
| 95 | c3s5pA_ |
|
not modelled |
8.9 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase b rpib from giardia2 lamblia
|
| 96 | d1p90a_ |
|
not modelled |
8.9 |
17 |
Fold:Ribonuclease H-like motif
Superfamily:Nitrogenase accessory factor-like
Family:Nitrogenase accessory factor |
| 97 | d1pdoa_ |
|
not modelled |
8.8 |
25 |
Fold:PTS system fructose IIA component-like
Superfamily:PTS system fructose IIA component-like
Family:EIIA-man component-like |
| 98 | c3mtqA_ |
|
not modelled |
8.6 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:putative phosphoenolpyruvate-dependent sugar
PDBTitle: crystal structure of a putative phosphoenolpyruvate-dependent sugar2 phosphotransferase system (pts) permease (kpn_04802) from klebsiella3 pneumoniae subsp. pneumoniae mgh 78578 at 1.70 a resolution
|
| 99 | d3pmga2 |
|
not modelled |
8.4 |
42 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |