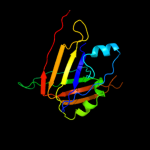
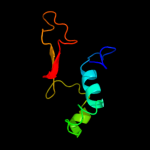
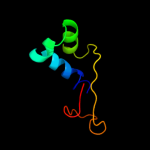
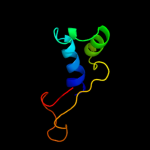
1 c3he1F_
100.0
28
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: major exported hcp3 protein;PDBTitle: secreted protein hcp3 from pseudomonas aeruginosa.
2 d1y12a1
100.0
17
Fold: Hcp1-likeSuperfamily: Hcp1-likeFamily: Hcp1-like3 c3v4hA_
100.0
17
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a type vi secretion system effector from yersinia2 pestis
4 c3eaaB_
100.0
10
PDB header: unknown functionChain: B: PDB Molecule: evpc;PDBTitle: structure of a type six secretion system protein
5 d2gykb1
99.7
32
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: HNH-motif6 d2jb0b1
97.8
31
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: HNH-motif7 c7ceiB_
97.7
28
PDB header: immune systemChain: B: PDB Molecule: protein (colicin e7 immunity protein);PDBTitle: the endonuclease domain of colicin e7 in complex with its inhibitor2 im7 protein
8 c2qgpA_
92.5
23
PDB header: hydrolaseChain: A: PDB Molecule: hnh endonuclease;PDBTitle: x-ray structure of the nhn endonuclease from geobacter2 metallireducens. northeast structural genomics consortium3 target gmr87.
9 c2joxA_
64.6
24
PDB header: transcriptionChain: A: PDB Molecule: churchill protein;PDBTitle: embryonic neural inducing factor churchill is not a dna-2 binding zinc finger protein: solution structure reveals a3 solvent-exposed beta-sheet and zinc binuclear cluster
10 d1n9pa_
59.7
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Cytoplasmic domain of inward rectifier potassium channel11 c1u3eM_
48.2
22
PDB header: dna binding protein/dnaChain: M: PDB Molecule: hnh homing endonuclease;PDBTitle: dna binding and cleavage by the hnh homing endonuclease i-hmui
12 c2gq1A_
43.5
26
PDB header: hydrolaseChain: A: PDB Molecule: fructose-1,6-bisphosphatase;PDBTitle: crystal structure of recombinant type i fructose-1,6-bisphosphatase2 from escherichia coli complexed with sulfate ions
13 d1xl4a1
41.6
16
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Cytoplasmic domain of inward rectifier potassium channel14 c1u4fD_
27.9
40
PDB header: allergenChain: D: PDB Molecule: inward rectifier potassium channel 2;PDBTitle: crystal structure of cytoplasmic domains of irk1 (kir2.1)2 channel
15 c2gixC_
27.3
43
PDB header: metal transportChain: C: PDB Molecule: inward rectifier potassium channel 2;PDBTitle: cytoplasmic domain structure of kir2.1 containing2 andersen's mutation r218q and rescue mutation t309k
16 d1bk4a_
27.0
25
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like17 d1nuwa_
26.8
23
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like18 c2xkyI_
26.0
43
PDB header: metal transportChain: I: PDB Molecule: inward rectifier potassium channel 2;PDBTitle: single particle analysis of kir2.1nc_4 in negative stain
19 c2e4fA_
25.0
12
PDB header: transport proteinChain: A: PDB Molecule: g protein-activated inward rectifier potassium channel 2;PDBTitle: crystal structure of the cytoplasmic domain of g-protein-gated inward2 rectifier potassium channel kir3.2
20 d1ftaa_
24.5
23
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like21 c2fhyL_
not modelled
24.2
23
PDB header: hydrolaseChain: L: PDB Molecule: fructose-1,6-bisphosphatase 1;PDBTitle: structure of human liver fpbase complexed with a novel2 benzoxazole as allosteric inhibitor
22 d2ouwa1
not modelled
22.8
13
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: TTHA0727-like23 d1p7ba1
not modelled
20.3
43
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Cytoplasmic domain of inward rectifier potassium channel24 d1u3em1
not modelled
18.5
17
Fold: His-Me finger endonucleasesSuperfamily: His-Me finger endonucleasesFamily: Intron-encoded homing endonucleases25 d1ne9a1
not modelled
18.3
17
Fold: Acyl-CoA N-acyltransferases (Nat)Superfamily: Acyl-CoA N-acyltransferases (Nat)Family: FemXAB nonribosomal peptidyltransferases26 c3jycA_
not modelled
15.8
17
PDB header: metal transportChain: A: PDB Molecule: inward-rectifier k+ channel kir2.2;PDBTitle: crystal structure of the eukaryotic strong inward-rectifier2 k+ channel kir2.2 at 3.1 angstrom resolution
27 d1skye2
not modelled
15.8
29
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: N-terminal domain of alpha and beta subunits of F1 ATP synthaseFamily: N-terminal domain of alpha and beta subunits of F1 ATP synthase28 c2qp2A_
not modelled
15.5
21
PDB header: unknown functionChain: A: PDB Molecule: unknown protein;PDBTitle: structure of a macpf/perforin-like protein
29 c3cfhB_
not modelled
14.9
18
PDB header: fluorescent proteinChain: B: PDB Molecule: gfp-like photoswitchable fluorescent protein;PDBTitle: photoswitchable red fluorescent protein psrfp, off-state
30 c2gw4D_
not modelled
14.8
21
PDB header: luminescent proteinChain: D: PDB Molecule: kaede;PDBTitle: crystal structure of stony coral fluorescent protein kaede, red form
31 c1zggA_
not modelled
13.7
12
PDB header: hydrolaseChain: A: PDB Molecule: putative low molecular weight protein-tyrosine-PDBTitle: solution structure of a low molecular weight protein2 tyrosine phosphatase from bacillus subtilis
32 c2otbB_
not modelled
13.5
21
PDB header: fluorescent proteinChain: B: PDB Molecule: gfp-like fluorescent chromoprotein cfp484;PDBTitle: crystal structure of a monomeric cyan fluorescent protein2 in the fluorescent state
33 c1p7bB_
not modelled
13.3
47
PDB header: metal transportChain: B: PDB Molecule: integral membrane channel and cytosolic domains;PDBTitle: crystal structure of an inward rectifier potassium channel
34 c3lf4B_
not modelled
13.3
21
PDB header: fluorescent proteinChain: B: PDB Molecule: fluorescent timer precursor blue102;PDBTitle: crystal structure of fluorescent timer precursor blue102
35 c2y7cA_
not modelled
12.7
25
PDB header: transferaseChain: A: PDB Molecule: type-1 restriction enzyme ecoki specificity protein;PDBTitle: atomic model of the ocr-bound methylase complex from the2 type i restriction-modification enzyme ecoki (m2s1). based3 on fitting into em map 1534.
36 d2fzta1
not modelled
12.3
26
Fold: Methionine synthase domain-likeSuperfamily: TM0693-likeFamily: TM0693-like37 c2zo6A_
not modelled
11.8
29
PDB header: luminescent proteinChain: A: PDB Molecule: cyan-emitting gfp-like protein, kusabira-cyan (kcy);PDBTitle: crystal structure of kusabira-cyan (kcy), a cyan-emitting gfp-like2 protein
38 d1xqma_
not modelled
11.4
18
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins39 c2g3dB_
not modelled
11.4
21
PDB header: luminescent proteinChain: B: PDB Molecule: green fluorescent protein;PDBTitle: structure of s65g y66a gfp variant after spontaneous2 peptide hydrolysis
40 c2ddcA_
not modelled
11.4
29
PDB header: luminescent proteinChain: A: PDB Molecule: photoconvertible fluorescent protein;PDBTitle: unique behavior of a histidine responsible for an2 engineered green-to-red photoconversion process
41 d1ggxa_
not modelled
11.1
21
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins42 d1d9qa_
not modelled
10.9
20
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like43 c3l4hA_
not modelled
10.9
16
PDB header: protein bindingChain: A: PDB Molecule: e3 ubiquitin-protein ligase hecw1;PDBTitle: helical box domain and second ww domain of the human e3 ubiquitin-2 protein ligase hecw1
44 c2zmwC_
not modelled
10.8
25
PDB header: luminescent proteinChain: C: PDB Molecule: fluorescent protein;PDBTitle: crystal structure of monomeric kusabira-orange (mko),2 orange-emitting gfp-like protein, at ph 6.0
45 d1spia_
not modelled
10.7
24
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like46 c2c9jG_
not modelled
10.7
25
PDB header: luminescent proteinChain: G: PDB Molecule: green fluorescent protein fp512;PDBTitle: structure of the fluorescent protein cmfp512 at 1.35a from2 cerianthus membranaceus
47 d1fc2c_
not modelled
10.4
22
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: Immunoglobulin-binding protein A modules48 c2zo7A_
not modelled
10.3
29
PDB header: luminescent proteinChain: A: PDB Molecule: cyan/green-emitting gfp-like protein, kusabira-cyan mutantPDBTitle: crystal structure of a kusabira-cyan mutant (kcy-r1), a cyan/green-2 emitting gfp-like protein
49 c1olpB_
not modelled
10.3
17
PDB header: hydrolaseChain: B: PDB Molecule: alpha-toxin;PDBTitle: alpha toxin from clostridium absonum
50 c2g9lA_
not modelled
10.2
38
PDB header: antibioticChain: A: PDB Molecule: gaegurin-4;PDBTitle: the high-resolution solution conformation of an2 antimicrobial peptide gaegurin 4 and its mode of membrane3 interaction
51 d2rh7a1
not modelled
10.0
21
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins52 c3cglE_
not modelled
10.0
21
PDB header: fluorescent proteinChain: E: PDB Molecule: gfp-like fluorescent chromoprotein dsfp483;PDBTitle: crystal structure and raman studies of dsfp483, a cyan fluorescent2 protein from discosoma striata
53 c1yzwB_
not modelled
9.9
18
PDB header: luminescent proteinChain: B: PDB Molecule: gfp-like non-fluorescent chromoprotein;PDBTitle: the 2.1a crystal structure of the far-red fluorescent2 protein hcred: inherent conformational flexibility of the3 chromophore
54 c2bjqA_
not modelled
9.8
15
PDB header: motilityChain: A: PDB Molecule: mfp2a;PDBTitle: crystal structure of the nematode sperm cell motility2 protein mfp2
55 c3gb3B_
not modelled
9.7
7
PDB header: fluorescent proteinChain: B: PDB Molecule: killerred;PDBTitle: x-ray structure of genetically encoded photosensitizer2 killerred in native form
56 c3simA_
not modelled
9.4
25
PDB header: hydrolaseChain: A: PDB Molecule: protein, family 18 chitinase;PDBTitle: crystallographic structure analysis of family 18 chitinase from crocus2 vernus
57 c2kxhB_
not modelled
9.0
64
PDB header: protein bindingChain: B: PDB Molecule: peptide of far upstream element-binding protein 1;PDBTitle: solution structure of the first two rrm domains of fir in the complex2 with fbp nbox peptide
58 c3myuB_
not modelled
8.8
20
PDB header: vib binding proteinChain: B: PDB Molecule: high affinity transport system protein p37;PDBTitle: mycoplasma genitalium mg289
59 c3sggA_
not modelled
8.8
36
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical hydrolase;PDBTitle: crystal structure of a hypothetical hydrolase (bt_2193) from2 bacteroides thetaiotaomicron vpi-5482 at 1.25 a resolution
60 c2y50A_
not modelled
8.7
23
PDB header: hydrolaseChain: A: PDB Molecule: collagenase;PDBTitle: crystal structure of collagenase g from clostridium2 histolyticum at 2.80 angstrom resolution
61 c2c9iG_
not modelled
8.7
18
PDB header: luminescent proteinChain: G: PDB Molecule: green fluorescent protein asfp499;PDBTitle: structure of the fluorescent protein asfp499 from anemonia2 sulcata
62 c2ib5H_
not modelled
8.7
14
PDB header: luminescent proteinChain: H: PDB Molecule: chromo protein;PDBTitle: structural characterization of a blue chromoprotein and its yellow2 mutant from the sea anemone cnidopus japonicus
63 c2z6zA_
not modelled
8.6
25
PDB header: fluorescent proteinChain: A: PDB Molecule: fluorescent protein dronpa;PDBTitle: crystal structure of a photoswitchable gfp-like protein2 dronpa in the bright-state
64 c2xfyA_
not modelled
8.5
21
PDB header: hydrolaseChain: A: PDB Molecule: beta-amylase;PDBTitle: crystal structure of barley beta-amylase complexed with2 alpha-cyclodextrin
65 d1moua_
not modelled
8.4
21
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins66 c2icrD_
not modelled
8.4
25
PDB header: fluorescent proteinChain: D: PDB Molecule: red fluorescent protein zoanrfp;PDBTitle: red fluorescent protein zrfp574 from zoanthus sp.
67 c2hpwA_
not modelled
8.3
21
PDB header: luminescent proteinChain: A: PDB Molecule: green fluorescent protein;PDBTitle: green fluorescent protein from clytia gregaria
68 c3eplA_
not modelled
8.3
17
PDB header: transferase/rnaChain: A: PDB Molecule: trna isopentenyltransferase;PDBTitle: crystallographic snapshots of eukaryotic2 dimethylallyltransferase acting on trna: insight into trna3 recognition and reaction mechanism
69 d1deeg_
not modelled
8.2
22
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: Immunoglobulin-binding protein A modules70 c1amlA_
not modelled
8.1
50
PDB header: serine protease inhibitorChain: A: PDB Molecule: amyloid a4;PDBTitle: the alzheimer`s disease amyloid a4 peptide (residues 1-40)
71 d2jwda1
not modelled
7.8
22
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: Immunoglobulin-binding protein A modules72 c3nezB_
not modelled
7.8
21
PDB header: fluorescent proteinChain: B: PDB Molecule: mrojoa;PDBTitle: mrojoa
73 d1uisa_
not modelled
7.7
18
Fold: GFP-likeSuperfamily: GFP-likeFamily: Fluorescent proteins74 d2cwqa1
not modelled
7.5
14
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: TTHA0727-like75 c3uksB_
not modelled
7.5
17
PDB header: hydrolaseChain: B: PDB Molecule: sedoheptulose-1,7 bisphosphatase, putative;PDBTitle: 1.85 angstrom crystal structure of putative sedoheptulose-1,72 bisphosphatase from toxoplasma gondii
76 d1hcra_
not modelled
7.1
63
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain77 d1ijwc_
not modelled
7.1
63
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Recombinase DNA-binding domain78 c2y6xA_
not modelled
7.0
19
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii 11 kd protein;PDBTitle: structure of psb27 from thermosynechococcus elongatus
79 c3t7hB_
not modelled
6.9
19
PDB header: ligaseChain: B: PDB Molecule: ubiquitin-like modifier-activating enzyme atg7;PDBTitle: atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and2 mechanism in the autophagy pathway
80 d1t3la2
not modelled
6.8
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases81 d1x9aa_
not modelled
6.7
22
Fold: DsrEFH-likeSuperfamily: DsrEFH-likeFamily: DsrH-like82 d1lrza1
not modelled
6.7
35
Fold: Long alpha-hairpinSuperfamily: tRNA-binding armFamily: Methicillin resistance protein FemA probable tRNA-binding arm83 d1khoa2
not modelled
6.6
21
Fold: Lipase/lipooxygenase domain (PLAT/LH2 domain)Superfamily: Lipase/lipooxygenase domain (PLAT/LH2 domain)Family: Alpha-toxin, C-terminal domain84 d1lp1b_
not modelled
6.6
22
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Bacterial immunoglobulin/albumin-binding domainsFamily: Immunoglobulin-binding protein A modules85 d1vyva2
not modelled
6.4
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases86 d1vyua2
not modelled
6.3
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases87 d1saza1
not modelled
6.2
29
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Acetokinase-like88 c2qksA_
not modelled
6.2
19
PDB header: metal transportChain: A: PDB Molecule: kir3.1-prokaryotic kir channel chimera;PDBTitle: crystal structure of a kir3.1-prokaryotic kir channel chimera
89 d2pstx1
not modelled
6.2
20
Fold: MbtH/L9 domain-likeSuperfamily: MbtH-likeFamily: MbtH-like90 c3nezA_
not modelled
6.2
16
PDB header: fluorescent proteinChain: A: PDB Molecule: mrojoa;PDBTitle: mrojoa
91 c3d3qB_
not modelled
6.0
12
PDB header: transferaseChain: B: PDB Molecule: trna delta(2)-isopentenylpyrophosphatePDBTitle: crystal structure of trna delta(2)-isopentenylpyrophosphate2 transferase (se0981) from staphylococcus epidermidis.3 northeast structural genomics consortium target ser100
92 c1y2iC_
not modelled
6.0
39
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein s0862;PDBTitle: crystal structure of mcsg target apc27401 from shigella2 flexneri
93 d1y2ia_
not modelled
6.0
39
Fold: Dodecin subunit-likeSuperfamily: YbjQ-likeFamily: YbjQ-like94 d1ogmx1
not modelled
6.0
28
Fold: Dextranase, N-terminal domainSuperfamily: Dextranase, N-terminal domainFamily: Dextranase, N-terminal domain95 c3akoG_
not modelled
6.0
24
PDB header: fluorescent proteinChain: G: PDB Molecule: venus;PDBTitle: crystal structure of the reassembled venus
96 c2yicC_
not modelled
5.7
12
PDB header: lyaseChain: C: PDB Molecule: 2-oxoglutarate decarboxylase;PDBTitle: crystal structure of the suca domain of mycobacterium smegmatis2 alpha-ketoglutarate decarboxylase (triclinic form)
97 c2okrC_
not modelled
5.6
38
PDB header: transferaseChain: C: PDB Molecule: map kinase-activated protein kinase 2;PDBTitle: crystal structure of the p38a-mapkap kinase 2 heterodimer
98 c2okrF_
not modelled
5.6
38
PDB header: transferaseChain: F: PDB Molecule: map kinase-activated protein kinase 2;PDBTitle: crystal structure of the p38a-mapkap kinase 2 heterodimer
99 c3nb0A_
not modelled
5.5
19
PDB header: transferaseChain: A: PDB Molecule: glycogen [starch] synthase isoform 2;PDBTitle: glucose-6-phosphate activated form of yeast glycogen synthase