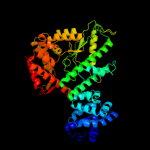
| 1 | c3k7dA_ |
|
 |
100.0 |
100 |
PDB header:transferase
Chain: A: PDB Molecule:glutamate-ammonia-ligase adenylyltransferase;
PDBTitle: c-terminal (adenylylation) domain of e.coli glutamine synthetase2 adenylyltransferase
|
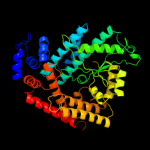
| 2 | c1v4aA_ |
|
 |
100.0 |
97 |
PDB header:transferase
Chain: A: PDB Molecule:glutamate-ammonia-ligase adenylyltransferase;
PDBTitle: structure of the n-terminal domain of escherichia coli2 glutamine synthetase adenylyltransferase
|
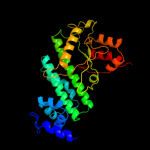
| 3 | d1v4aa2 |
|
 |
100.0 |
97 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:GlnE-like domain |
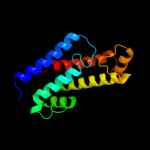
| 4 | d1v4aa1 |
|
 |
100.0 |
97 |
Fold:Four-helical up-and-down bundle
Superfamily:Nucleotidyltransferase substrate binding subunit/domain
Family:Glutamine synthase adenylyltransferase GlnE, domain 2 |
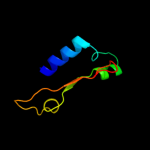
| 5 | d1knya2 |
|
 |
95.7 |
22 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Kanamycin nucleotidyltransferase (KNTase), N-terminal domain |
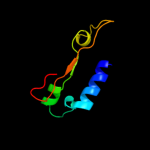
| 6 | c1knyA_ |
|
 |
95.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:kanamycin nucleotidyltransferase;
PDBTitle: kanamycin nucleotidyltransferase
|
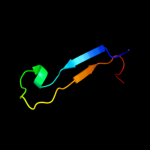
| 7 | c2rffA_ |
|
 |
93.7 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:putative nucleotidyltransferase;
PDBTitle: crystal structure of a putative nucleotidyltransferase2 (np_343093.1) from sulfolobus solfataricus at 1.40 a3 resolution
|
| 8 | d1no5a_ |
|
 |
91.0 |
26 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Catalytic subunit of bi-partite nucleotidyltransferase |
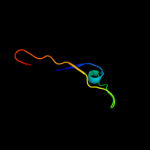
| 9 | d1wota_ |
|
 |
89.1 |
31 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Catalytic subunit of bi-partite nucleotidyltransferase |
| 10 | c3c66B_ |
|
 |
87.3 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:poly(a) polymerase;
PDBTitle: yeast poly(a) polymerase in complex with fip1 residues 80-105
|
| 11 | c1q78A_ |
|
 |
86.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:poly(a) polymerase alpha;
PDBTitle: crystal structure of poly(a) polymerase in complex with 3'-2 datp and magnesium chloride
|
| 12 | d1ylqa1 |
|
 |
85.1 |
27 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Catalytic subunit of bi-partite nucleotidyltransferase |
| 13 | c3jz0B_ |
|
 |
76.8 |
20 |
PDB header:transferase/antibiotic
Chain: B: PDB Molecule:lincosamide nucleotidyltransferase;
PDBTitle: linb complexed with clindamycin and ampcpp
|
| 14 | c3o7kA_ |
|
 |
69.4 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:ohcu decarboxylase;
PDBTitle: crystal structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline2 decarboxylase from klebsiella pneumoniae
|
| 15 | d1q79a2 |
|
 |
66.9 |
14 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Poly(A) polymerase, PAP, N-terminal domain |
| 16 | c1kdhA_ |
|
 |
60.8 |
13 |
PDB header:transferase/dna
Chain: A: PDB Molecule:terminal deoxynucleotidyltransferase short
PDBTitle: binary complex of murine terminal deoxynucleotidyl2 transferase with a primer single stranded dna
|
| 17 | d2o70a1 |
|
 |
57.8 |
18 |
Fold:UraD-like
Superfamily:UraD-Like
Family:UraD-like |
| 18 | d2q66a2 |
|
 |
57.2 |
12 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Poly(A) polymerase, PAP, N-terminal domain |
| 19 | c3hvzB_ |
|
 |
55.9 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the tgs domain of the clolep_03100 protein from2 clostridium leptum, northeast structural genomics consortium target3 qlr13a
|
| 20 | c1sz1A_ |
|
 |
54.5 |
20 |
PDB header:transferase/rna
Chain: A: PDB Molecule:trna nucleotidyltransferase;
PDBTitle: mechanism of cca-adding enzymes specificity revealed by crystal2 structures of ternary complexes
|
| 21 | d1r89a2 |
|
not modelled |
49.5 |
18 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Archaeal tRNA CCA-adding enzyme catalytic domain |
| 22 | d2fmpa3 |
|
not modelled |
47.1 |
10 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
| 23 | d1d5ya2 |
|
not modelled |
47.1 |
23 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:AraC type transcriptional activator |
| 24 | c3r66A_ |
|
not modelled |
44.0 |
13 |
PDB header:viral protein/antiviral protein
Chain: A: PDB Molecule:non-structural protein 1;
PDBTitle: crystal structure of human isg15 in complex with ns1 n-terminal region2 from influenza virus b, northeast structural genomics consortium3 target ids hx6481, hr2873, and or2
|
| 25 | d2bcqa3 |
|
not modelled |
43.9 |
17 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
| 26 | d2vana2 |
|
not modelled |
42.0 |
13 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
| 27 | d1xeqa1 |
|
not modelled |
39.7 |
13 |
Fold:S15/NS1 RNA-binding domain
Superfamily:S15/NS1 RNA-binding domain
Family:N-terminal, RNA-binding domain of nonstructural protein NS1 |
| 28 | d1jqra_ |
|
not modelled |
39.6 |
33 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
| 29 | c2kmmA_ |
|
not modelled |
36.4 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:guanosine-3',5'-bis(diphosphate) 3'-
PDBTitle: solution nmr structure of the tgs domain of pg1808 from2 porphyromonas gingivalis. northeast structural genomics3 consortium target pgr122a (418-481)
|
| 30 | d1bl0a2 |
|
not modelled |
34.8 |
26 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:AraC type transcriptional activator |
| 31 | d1phna_ |
|
not modelled |
33.1 |
16 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
| 32 | c2q0dA_ |
|
not modelled |
30.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:rna uridylyl transferase;
PDBTitle: terminal uridylyl transferase 4 from trypanosoma brucei2 with bound atp
|
| 33 | d1jmsa4 |
|
not modelled |
29.8 |
14 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
| 34 | c3ng3A_ |
|
not modelled |
29.5 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of deoxyribose phosphate aldolase from mycobacterium2 avium 104 in a schiff base with an unknown aldehyde
|
| 35 | d1vi7a1 |
|
not modelled |
28.1 |
35 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:YigZ N-terminal domain-like |
| 36 | c2dt5A_ |
|
not modelled |
25.2 |
28 |
PDB header:dna binding protein
Chain: A: PDB Molecule:at-rich dna-binding protein;
PDBTitle: crystal structure of ttha1657 (at-rich dna-binding protein) from2 thermus thermophilus hb8
|
| 37 | c1nomA_ |
|
not modelled |
24.6 |
14 |
PDB header:nucleotidyltransferase
Chain: A: PDB Molecule:dna polymerase beta;
PDBTitle: dna polymerase beta (pol b) (e.c.2.7.7.7), 31-kd domain; soaked in the2 presence of mncl2 (5 millimolar)
|
| 38 | d2dt5a2 |
|
not modelled |
23.8 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Transcriptional repressor Rex, C-terminal domain |
| 39 | d2cvea1 |
|
not modelled |
23.6 |
44 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:YigZ N-terminal domain-like |
| 40 | c3f2eA_ |
|
not modelled |
23.3 |
19 |
PDB header:viral protein
Chain: A: PDB Molecule:sirv coat protein;
PDBTitle: crystal structure of yellowstone sirv coat protein c-2 terminus
|
| 41 | d2b8ta1 |
|
not modelled |
22.4 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Type II thymidine kinase |
| 42 | d1o0ya_ |
|
not modelled |
22.4 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 43 | c3hiyA_ |
|
not modelled |
21.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:minor editosome-associated tutase;
PDBTitle: minor editosome-associated tutase 1 with bound utp and mg
|
| 44 | d1cpca_ |
|
not modelled |
21.4 |
14 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
| 45 | d1qmga2 |
|
not modelled |
20.6 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 46 | c3ngjC_ |
|
not modelled |
20.4 |
22 |
PDB header:lyase
Chain: C: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 entamoeba histolytica
|
| 47 | c3nybA_ |
|
not modelled |
18.2 |
11 |
PDB header:transferase/rna binding protein
Chain: A: PDB Molecule:poly(a) rna polymerase protein 2;
PDBTitle: structure and function of the polymerase core of tramp, a rna2 surveillance complex
|
| 48 | c2ihmA_ |
|
not modelled |
18.1 |
14 |
PDB header:transferase/dna
Chain: A: PDB Molecule:dna polymerase mu;
PDBTitle: polymerase mu in ternary complex with gapped 11mer dna2 duplex and bound incoming nucleotide
|
| 49 | c2b8tA_ |
|
not modelled |
17.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:thymidine kinase;
PDBTitle: crystal structure of thymidine kinase from u.urealyticum in2 complex with thymidine
|
| 50 | d1pvea_ |
|
not modelled |
17.2 |
13 |
Fold:XPC-binding domain
Superfamily:XPC-binding domain
Family:XPC-binding domain |
| 51 | d1r8ka_ |
|
not modelled |
16.6 |
23 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
| 52 | d1sfla_ |
|
not modelled |
16.5 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 53 | c8iczA_ |
|
not modelled |
16.1 |
11 |
PDB header:transferase/dna
Chain: A: PDB Molecule:protein (dna polymerase beta (e.c.2.7.7.7));
PDBTitle: dna polymerase beta (pol b) (e.c.2.7.7.7) complexed with2 seven base pairs of dna; soaked in the presence of of datp3 (1 millimolar), mncl2 (5 millimolar), and lithium sulfate4 (75 millimolar)
|
| 54 | c2bcuA_ |
|
not modelled |
15.7 |
22 |
PDB header:transferase, lyase/dna
Chain: A: PDB Molecule:dna polymerase lambda;
PDBTitle: dna polymerase lambda in complex with a dna duplex2 containing an unpaired damp and a t:t mismatch
|
| 55 | c2a4aB_ |
|
not modelled |
15.5 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: deoxyribose-phosphate aldolase from p. yoelii
|
| 56 | c1ug9A_ |
|
not modelled |
15.3 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:glucodextranase;
PDBTitle: crystal structure of glucodextranase from arthrobacter globiformis i42
|
| 57 | c2xvcA_ |
|
not modelled |
15.3 |
38 |
PDB header:cell cycle
Chain: A: PDB Molecule:escrt-iii;
PDBTitle: molecular and structural basis of escrt-iii recruitment to2 membranes during archaeal cell division
|
| 58 | c2jq5A_ |
|
not modelled |
14.9 |
17 |
PDB header:structural genomics
Chain: A: PDB Molecule:sec-c motif;
PDBTitle: solution structure of rpa3114, a sec-c motif containing2 protein from rhodopseudomonas palustris; northeast3 structural genomics consortium target rpt5 / ontario4 center for structural proteomics target rp3097
|
| 59 | c1tltB_ |
|
not modelled |
14.9 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase (virulence factor mvim homolog);
PDBTitle: crystal structure of a putative oxidoreductase (virulence factor mvim2 homolog)
|
| 60 | d1ha7a_ |
|
not modelled |
14.8 |
13 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
| 61 | d1qt1a_ |
|
not modelled |
14.6 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
| 62 | c2j96B_ |
|
not modelled |
14.5 |
13 |
PDB header:photosynthesis
Chain: B: PDB Molecule:phycoerythrocyanin alpha chain;
PDBTitle: the e-configuration of alfa-phycoerythrocyanin
|
| 63 | d1mzha_ |
|
not modelled |
14.4 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 64 | c1yybA_ |
|
not modelled |
13.3 |
24 |
PDB header:apoptosis
Chain: A: PDB Molecule:programmed cell death protein 5;
PDBTitle: solution structure of 1-26 fragment of human programmed2 cell death 5 protein
|
| 65 | c3l9cA_ |
|
not modelled |
13.1 |
31 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: the crystal structure of smu.777 from streptococcus mutans ua159
|
| 66 | c3pq1A_ |
|
not modelled |
13.1 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:poly(a) rna polymerase;
PDBTitle: crystal structure of human mitochondrial poly(a) polymerase (papd1)
|
| 67 | c2gf5A_ |
|
not modelled |
13.0 |
14 |
PDB header:apoptosis
Chain: A: PDB Molecule:fadd protein;
PDBTitle: structure of intact fadd (mort1)
|
| 68 | c2oczA_ |
|
not modelled |
12.9 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: the structure of a putative 3-dehydroquinate dehydratase from2 streptococcus pyogenes.
|
| 69 | c1vh6A_ |
|
not modelled |
12.8 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:flagellar protein flis;
PDBTitle: crystal structure of a flagellar protein
|
| 70 | d1vh6a_ |
|
not modelled |
12.8 |
10 |
Fold:Four-helical up-and-down bundle
Superfamily:Flagellar export chaperone FliS
Family:Flagellar export chaperone FliS |
| 71 | c3oa3A_ |
|
not modelled |
12.7 |
26 |
PDB header:lyase
Chain: A: PDB Molecule:aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 coccidioides immitis
|
| 72 | d1oqya3 |
|
not modelled |
12.7 |
17 |
Fold:XPC-binding domain
Superfamily:XPC-binding domain
Family:XPC-binding domain |
| 73 | c1yveK_ |
|
not modelled |
12.6 |
23 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:acetohydroxy acid isomeroreductase;
PDBTitle: acetohydroxy acid isomeroreductase complexed with nadph,2 magnesium and inhibitor ipoha (n-hydroxy-n-3 isopropyloxamate)
|
| 74 | c2g5xA_ |
|
not modelled |
12.5 |
3 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribosome-inactivating protein;
PDBTitle: crystal structure of lychnin a type 1 ribosome inactivating2 protein (rip)
|
| 75 | d1sc6a1 |
|
not modelled |
12.5 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 76 | c1wr1B_ |
|
not modelled |
12.4 |
22 |
PDB header:signaling protein
Chain: B: PDB Molecule:ubiquitin-like protein dsk2;
PDBTitle: the complex sturcture of dsk2p uba with ubiquitin
|
| 77 | c2of5A_ |
|
not modelled |
12.4 |
17 |
PDB header:apoptosis
Chain: A: PDB Molecule:death domain-containing protein cradd;
PDBTitle: oligomeric death domain complex
|
| 78 | c1wxpA_ |
|
not modelled |
12.3 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:tho complex subunit 1;
PDBTitle: solution structure of the death domain of nuclear matrix2 protein p84
|
| 79 | d1gqna_ |
|
not modelled |
12.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 80 | c1f7uA_ |
|
not modelled |
11.8 |
19 |
PDB header:ligase/rna
Chain: A: PDB Molecule:arginyl-trna synthetase;
PDBTitle: crystal structure of the arginyl-trna synthetase complexed with the2 trna(arg) and l-arg
|
| 81 | d2fi0a1 |
|
not modelled |
11.8 |
17 |
Fold:SP0561-like
Superfamily:SP0561-like
Family:SP0561-like |
| 82 | c3iwfA_ |
|
not modelled |
11.7 |
15 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcription regulator rpir family;
PDBTitle: the crystal structure of the n-terminal domain of a rpir2 transcriptional regulator from staphylococcus epidermidis to 1.4a
|
| 83 | c1hlvA_ |
|
not modelled |
11.7 |
14 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:major centromere autoantigen b;
PDBTitle: crystal structure of cenp-b(1-129) complexed with the cenp-2 b box dna
|
| 84 | d2f4mb1 |
|
not modelled |
11.6 |
13 |
Fold:XPC-binding domain
Superfamily:XPC-binding domain
Family:XPC-binding domain |
| 85 | d1qm4a2 |
|
not modelled |
11.6 |
18 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 86 | d1p1xa_ |
|
not modelled |
11.5 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
| 87 | d1hlva2 |
|
not modelled |
11.4 |
14 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Centromere-binding |
| 88 | d1qp8a1 |
|
not modelled |
11.4 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 89 | c1x3wB_ |
|
not modelled |
11.3 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:uv excision repair protein rad23;
PDBTitle: structure of a peptide:n-glycanase-rad23 complex
|
| 90 | d1np3a2 |
|
not modelled |
11.2 |
43 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
| 91 | d1kn1a_ |
|
not modelled |
11.2 |
5 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
| 92 | c2ytuA_ |
|
not modelled |
11.1 |
15 |
PDB header:signaling protein
Chain: A: PDB Molecule:friend leukemia integration 1 transcription
PDBTitle: solution structure of the sam_pnt-domain of the human2 friend leukemiaintegration 1 transcription factor
|
| 93 | d1jboa_ |
|
not modelled |
11.0 |
17 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
| 94 | d1mxaa2 |
|
not modelled |
10.9 |
24 |
Fold:S-adenosylmethionine synthetase
Superfamily:S-adenosylmethionine synthetase
Family:S-adenosylmethionine synthetase |
| 95 | d1dxya1 |
|
not modelled |
10.6 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Formate/glycerate dehydrogenases, NAD-domain |
| 96 | c2cveA_ |
|
not modelled |
10.6 |
75 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ttha1053;
PDBTitle: crystal structure of a conserved hypothetical protein tt1547 from2 thermus thermophilus hb8
|
| 97 | c2vq3B_ |
|
not modelled |
10.6 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:metalloreductase steap3;
PDBTitle: crystal structure of the membrane proximal oxidoreductase2 domain of human steap3, the dominant ferric reductase of3 the erythroid transferrin cycle
|
| 98 | c3d19E_ |
|
not modelled |
10.4 |
16 |
PDB header:structural genomics, unknown function
Chain: E: PDB Molecule:conserved metalloprotein;
PDBTitle: crystal structure of a conserved metalloprotein from bacillus cereus
|
| 99 | c3l2iB_ |
|
not modelled |
10.4 |
25 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
|

